Comparing HITEMP and ExoMol¶
from exojax.spec import xsection
from exojax.spec.hitran import SijT, doppler_sigma, gamma_hitran, gamma_natural
from exojax.spec.exomol import gamma_exomol
from exojax.spec import moldb
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
plt.style.use('bmh')
First of all, set a wavenumber bin in the unit of wavenumber (cm-1). Here we set the wavenumber range as \(1000 \le \nu \le 10000\) (1/cm) with the resolution of 0.01 (1/cm).
We call moldb instance with the path of par file. If the par file does not exist, moldb will try to download it from HITRAN website.
# Setting wavenumber bins and loading HITEMP database
wav=np.linspace(22930.0,23000.0,4000,dtype=np.float64) #AA
nus=1.e8/wav[::-1] #cm-1
mdbCO_HITEMP=moldb.MdbHit('~/exojax/data/CO/05_HITEMP2019.par.bz2',nus)
bunziping
emf='~/exojax/data/CO/12C-16O/Li2015'
mdbCO_Li2015=moldb.MdbExomol(emf,nus)
Background atmosphere: H2
Reading transition file
Broadening code level= a0
default broadening parameters are used for 71 J lower states in 152 states
Define molecular weight of CO (\(\sim 12+16=28\)), temperature (K), and pressure (bar). Also, we here assume the 100 % CO atmosphere, i.e. the partial pressure = pressure.
from exojax.spec import molinfo
Mmol=molinfo.molmass("CO") # molecular weight
Tfix=1300.0 # we assume T=1300K
Pfix=0.99 # we compute P=1 bar=0.99+0.1
Ppart=0.01 #partial pressure of CO. here we assume a 1% CO atmosphere (very few).
partition function ratio \(q(T)\) is defined by
\(q(T) = Q(T)/Q(T_{ref})\); \(T_{ref}\)=296 K
Here, we use the partition function from HAPI
mdbCO_HITEMP.ExomolQT(emf) #use Q(T) from Exomol/Li2015
qt_HITEMP=mdbCO_HITEMP.qr_interp(Tfix)
qt_Li2015=mdbCO_Li2015.qr_interp(Tfix)
Let us compute the line strength S(T) at temperature of Tfix.
\(S (T;s_0,\nu_0,E_l,q(T)) = S_0 \frac{Q(T_{ref})}{Q(T)} \frac{e^{- h c E_l /k_B T}}{e^{- h c E_l /k_B T_{ref}}} \frac{1- e^{- h c \nu /k_B T}}{1-e^{- h c \nu /k_B T_{ref}}}= q_r(T)^{-1} e^{ s_0 - c_2 E_l (T^{-1} - T_{ref}^{-1})} \frac{1- e^{- c_2 \nu_0/ T}}{1-e^{- c_2 \nu_0/T_{ref}}}\)
\(s_0=\log_{e} S_0\) : logsij0
\(\nu_0\): nu_lines
\(E_l\) : elower
Why the input is \(s_0 = \log_{e} S_0\) instead of \(S_0\) in SijT? This is because the direct value of \(S_0\) is quite small and we need to use float32 for jax.
Sij_HITEMP=SijT(Tfix,mdbCO_HITEMP.logsij0,mdbCO_HITEMP.nu_lines,\
mdbCO_HITEMP.elower,qt_HITEMP)
Sij_Li2015=SijT(Tfix,mdbCO_Li2015.logsij0,mdbCO_Li2015.nu_lines,\
mdbCO_Li2015.elower,qt_Li2015)
Then, compute the Lorentz gamma factor (pressure+natural broadening)
\(\gamma_L = \gamma^p_L + \gamma^n_L\)
where the pressure broadning (HITEMP)
\(\gamma^p_L = (T/296K)^{-n_{air}} [ \alpha_{air} ( P - P_{part})/P_{atm} + \alpha_{self} P_{part}/P_{atm}]\)
\(P_{atm}\): 1 atm in the unit of bar (i.e. = 1.01325)
or
the pressure broadning (ExoMol)
\(\gamma^p_L = \alpha_{ref} ( T/T_{ref})^{-n_{texp}} ( P/P_{ref}),\)
and the natural broadening
\(\gamma^n_L = \frac{A}{4 \pi c}\)
gammaL_HITEMP = gamma_hitran(Pfix,Tfix, Ppart, mdbCO_HITEMP.n_air, \
mdbCO_HITEMP.gamma_air, mdbCO_HITEMP.gamma_self) \
+ gamma_natural(mdbCO_HITEMP.A)
gammaL_Li2015 = gamma_exomol(Pfix,Tfix,mdbCO_Li2015.n_Texp,mdbCO_Li2015.alpha_ref)\
+ gamma_natural(mdbCO_Li2015.A)
Thermal broadening
\(\sigma_D^{t} = \sqrt{\frac{k_B T}{M m_u}} \frac{\nu_0}{c}\)
# thermal doppler sigma
sigmaD_HITEMP=doppler_sigma(mdbCO_HITEMP.nu_lines,Tfix,Mmol)
sigmaD_Li2015=doppler_sigma(mdbCO_Li2015.nu_lines,Tfix,Mmol)
Then, the line center…
In HITRAN database, a slight pressure shift can be included using \(\delta_{air}\): \(\nu_0(P) = \nu_0 + \delta_{air} P\). But this shift is quite a bit.
#line center
nu0_HITEMP=mdbCO_HITEMP.nu_lines
nu0_Li2015=mdbCO_Li2015.nu_lines
Although it depends on your GPU, you might need to devide the computation into multiple loops because of the limitation of the GPU memory. Here we assume 30MB for GPU memory (not exactly, memory size for numatrix).
xsv_HITEMP=xsection(nus,nu0_HITEMP,sigmaD_HITEMP,gammaL_HITEMP\
,Sij_HITEMP,memory_size=30) #use 30MB GPU MEMORY for numax
xsv_Li2015=xsection(nus,nu0_Li2015,sigmaD_Li2015,gammaL_Li2015\
,Sij_Li2015,memory_size=30) #use 30MB GPU MEMORY for numax
100%|██████████| 12/12 [00:08<00:00, 1.31it/s]
100%|██████████| 1/1 [00:00<00:00, 1.08it/s]
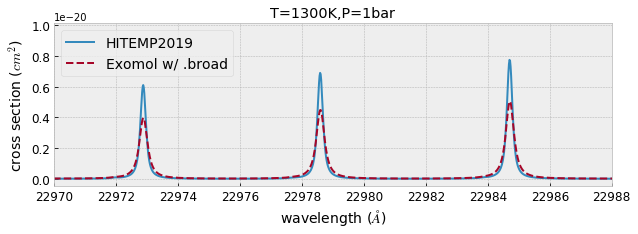
fig=plt.figure(figsize=(10,3))
ax=fig.add_subplot(111)
plt.plot(wav[::-1],xsv_HITEMP,lw=2,label="HITEMP2019")
plt.plot(wav[::-1],xsv_Li2015,lw=2,ls="dashed",label="Exomol w/ .broad")
plt.xlim(22970,22988)
plt.xlabel("wavelength ($\AA$)",fontsize=14)
plt.ylabel("cross section ($cm^{2}$)",fontsize=14)
plt.legend(loc="upper left",fontsize=14)
plt.tick_params(labelsize=12)
plt.savefig("co_comparison.pdf", bbox_inches="tight", pad_inches=0.0)
plt.savefig("co_comparison.png", bbox_inches="tight", pad_inches=0.0)
plt.title("T=1300K,P=1bar")
plt.show()
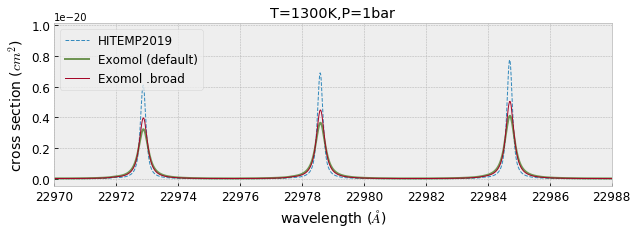
Here, we test to use the default broadening parameters in def file of exomol
mdbCO_Li2015.set_broadening(broadf=False) #use default broadning
gammaL_Li2015_def = gamma_exomol(Pfix,Tfix,mdbCO_Li2015.n_Texp,mdbCO_Li2015.alpha_ref)\
+ gamma_natural(mdbCO_Li2015.A)
xsv_Li2015_def=xsection(nus,nu0_Li2015,sigmaD_Li2015,gammaL_Li2015_def\
,Sij_Li2015,memory_size=30) #use 30MB GPU MEMORY for numax
100%|██████████| 1/1 [00:00<00:00, 20.04it/s]
No .broad file is given.
Plot it!
fig=plt.figure(figsize=(10,3))
ax=fig.add_subplot(111)
plt.plot(wav[::-1],xsv_HITEMP,lw=1,ls="dashed",label="HITEMP2019")
plt.plot(wav[::-1],xsv_Li2015_def,color="C3",lw=2,alpha=0.75,label="Exomol (default)")
plt.plot(wav[::-1],xsv_Li2015,lw=1,label="Exomol .broad")
plt.xlim(22970,22988)
plt.xlabel("wavelength ($\AA$)",fontsize=14)
plt.ylabel("cross section ($cm^{2}$)",fontsize=14)
plt.legend(loc="upper left",fontsize=12)
plt.tick_params(labelsize=12)
plt.savefig("co_comparison.pdf", bbox_inches="tight", pad_inches=0.0)
plt.savefig("co_comparison.png", bbox_inches="tight", pad_inches=0.0)
plt.title("T=1300K,P=1bar")
plt.show()
np.min(gammaL_HITEMP),np.max(gammaL_HITEMP)
(DeviceArray(0.01456154, dtype=float32),
DeviceArray(0.02531094, dtype=float32))
np.min(gammaL_Li2015),np.max(gammaL_Li2015)
(DeviceArray(0.02603155, dtype=float32),
DeviceArray(0.03306796, dtype=float32))
np.min(gammaL_Li2015_def),np.max(gammaL_Li2015_def)
(DeviceArray(0.03306796, dtype=float32),
DeviceArray(0.03306796, dtype=float32))