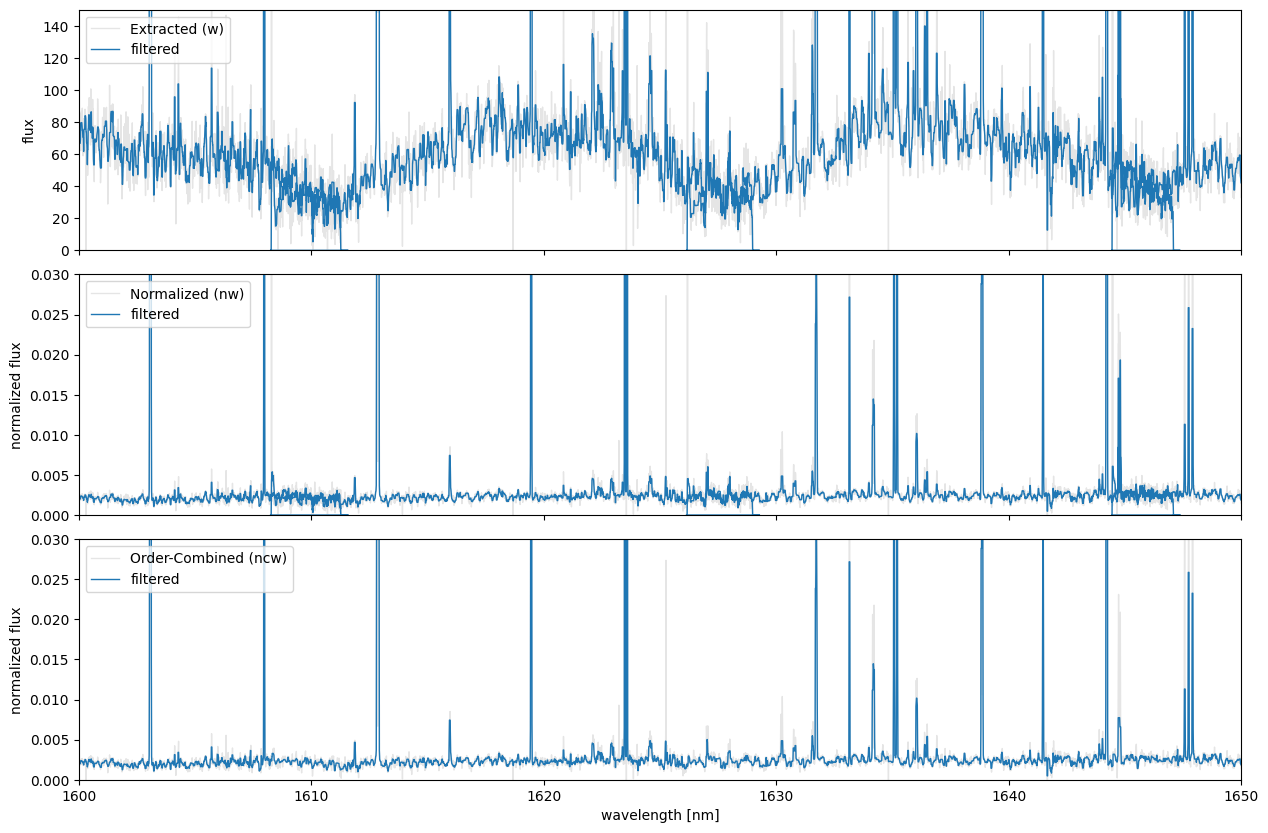
Output Spectra
When running :doc:./IRD_stream, three types of 1D spectra are
generated:
Wavelength calibrated 1D spectrum (**’w…_m?.dat’**) with columns:
$1: Wavelength [nm],$2: Order,$3: Counts.Normalized 1D spectrum (**’nw…_m?.dat’**) with columns:
$1: Wavelength [nm],$2: Order,$3: Counts,$4: S/N,$5: UncertaintiesOrder-combined normalized 1D spectrum (**’ncw…_m?.dat’**) with columns:
$1: Wavelength [nm],$2: Counts,$3: S/N,$4: Uncertainties
- The normalized spectrum (nw) is obtained by dividing the extracted spectrum (w) by the blaze function (`wblaze_?_m?.dat`).
- The order-combined spectrum (ncw) demonstrates an improved S/N in the wavelength region where adjacent orders overlap.
import pathlib
import pandas as pd
from scipy.signal import medfilt
import matplotlib.pyplot as plt
basedir = pathlib.Path('~/pyird/data/20210317/').expanduser()
anadir = basedir/'reduc/'
fitsid_target = [41510]
band = "h"
mmf = "mmf2"
if band=="h" and fitsid_target[0]%2==0:
fitsid_target = [x+1 for x in fitsid_target]
readargs = {"header": None, "sep": "\s+"}
names = ["wavelength [nm]", "order", "counts", "sn_ratio", "uncertainty"]
<>:10: SyntaxWarning: invalid escape sequence 's'
<>:10: SyntaxWarning: invalid escape sequence 's'
/var/folders/sb/rxwbk6kd4gldqpdvzxnhq4xm0000gn/T/ipykernel_11979/3404868316.py:10: SyntaxWarning: invalid escape sequence 's'
readargs = {"header": None, "sep": "s+"}
The figure shows the target spectra extracted using PyIRD.
- The sample dataset corresponds to the brown dwarf G196-3B.
- The emission-like signals observed in the spectra are likely due to sky (airglow) emissions or hotpixels that were not masked.
for fitsid in fitsid_target:
# Extracted Spectrum:
# $1: wavelength [nm], $2: order, $3: counts
wfile_path = anadir / f"w{fitsid}_{mmf[0]}{mmf[-1]}.dat"
wspec = pd.read_csv(wfile_path, names = names[:3], **readargs)
# Normalized Spectrum:
# $1: wavelength [nm], $2: order, $3: counts, $4: sn_ratio, $5: uncertainty
nwfile_path = anadir / f"nw{fitsid}_{mmf[0]}{mmf[-1]}.dat"
nwspec = pd.read_csv(nwfile_path, names = names, **readargs)
# Order-combined Spectrum:
# $1: wavelength [nm], $2: counts, $3: sn_ratio, $4: uncertainty
ncwfile_path = anadir / f"ncw{fitsid}_{mmf[0]}{mmf[-1]}.dat"
names_ncw = [x for i, x in enumerate(names) if i != 1]
ncwspec = pd.read_csv(ncwfile_path, names = names_ncw, **readargs)
# Plot
fig, axs = plt.subplots(3, 1, figsize=(15,10), sharex=True)
plt.subplots_adjust(hspace=0.1)
labels = ["Extracted (w)", "Normalized (nw)", "Order-Combined (ncw)"]
for i, spec in enumerate([wspec, nwspec, ncwspec]):
axs[i].plot(spec["wavelength [nm]"], spec["counts"], lw=1, color="grey", alpha=0.2, label=labels[i])
axs[i].plot(spec["wavelength [nm]"], medfilt(spec["counts"], 5), lw=1, label="filtered")
for ax in axs:
ax.legend(loc="upper left")
axs[0].set(xlim=(1600, 1650),#(wspec["wavelength [nm]"].min(),wspec["wavelength [nm]"].max()),
ylim=(0, 150),
ylabel="flux")
axs[1].set(ylim=(0,0.03),
ylabel="normalized flux")
axs[2].set(ylim=(0,0.03),
xlabel="wavelength [nm]",
ylabel="normalized flux")
plt.show()