Handling Apertures
from pyird.utils import irdstream
import pathlib
Single Fiber Mode (nap = 21 or nap = 51)
In some observations, only the star fiber (mmf2) is used, without
the comb (or speckle) fiber (mmf1). In such cases, extract the
desired apertures using aptrace with nap = 21 for H-band images
or nap = 51 for Y/J-band images.
#--------SETTINGS--------#
basedir = pathlib.Path('~/pyird/data/20210317/').expanduser()
band = 'h' #'h' or 'y'
mmf = 'mmf2' #'mmf1' (comb fiber) or 'mmf2' (star fiber)
readout_noise_mode = "default" #'real' or 'default'
datadir_flat = basedir/'flat/'
anadir = basedir/'reduc/'
fitsid_flat_star = list(range(41804,41904,2))
#-------------------------#
## FLAT_STAR
flat_star=irdstream.Stream2D("flat_star",datadir_flat,anadir)
flat_star.fitsid=fitsid_flat_star.copy()
# aperture extraction
flat_star.band=band
print(flat_star.band,' band')
if band=='h' and flat_star.fitsid[0]%2==0:
flat_star.fitsid_increment()
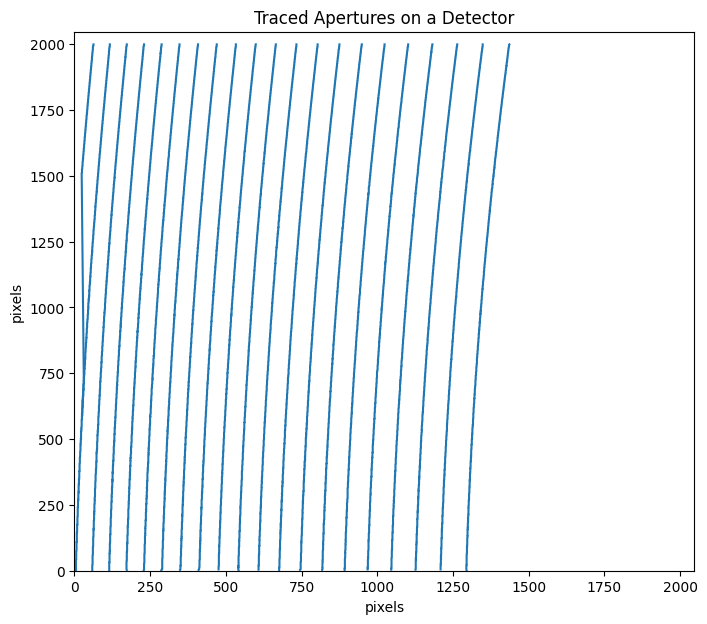
trace_mmf=flat_star.aptrace(cutrow = 1500,nap=21)
elif band=='y':
trace_mmf=flat_star.aptrace(cutrow = 1000,nap=51)
No fitsid yet.
h band
fitsid incremented: [41805, 41807, 41809, 41811, 41813, 41815, 41817, 41819, 41821, 41823, 41825, 41827, 41829, 41831, 41833, 41835, 41837, 41839, 41841, 41843, 41845, 41847, 41849, 41851, 41853, 41855, 41857, 41859, 41861, 41863, 41865, 41867, 41869, 41871, 41873, 41875, 41877, 41879, 41881, 41883, 41885, 41887, 41889, 41891, 41893, 41895, 41897, 41899, 41901, 41903]
median combine:
100%|█████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 50/50 [00:00<00:00, 1032.06it/s]
/Users/yuikasagi/git/pyird/src/pyird/image/aptrace.py:251: UserWarning: Looks a single fiber aperture on the detector.
warnings.warn("Looks a single fiber aperture on the detector.", UserWarning)
cross-section: row 1502
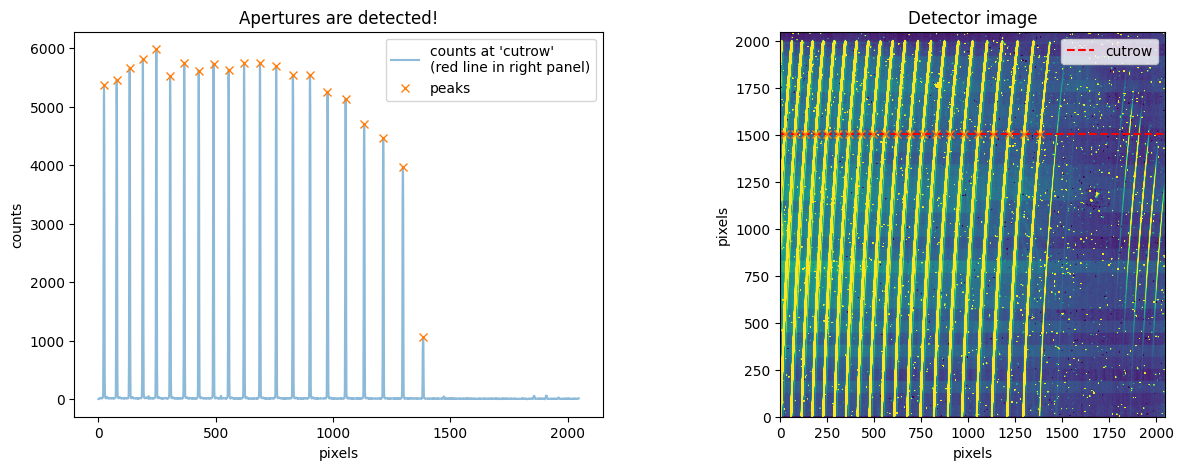
100%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 21/21 [00:03<00:00, 6.35it/s]
Wavelength Calibration with User Defined Apertures (nap < 20 for H or nap < 51 for YJ)
If you want to extract apertures of your choice, you can use aptrace
with an arbitrary value for nap.
NOTE: Currently, only single fiber mode is supported. The use of both fibers simultaneously is not accounted for.
Set ign_ord to specify the aperture orders you do not want to trace.
Ensure that the condition len(ign_ord) + nap = 21 for H-band images
or len(ign_ord) + nap = 51 for Y/J-band images is satisfied.
## FLAT_STAR
flat_star_apcut=irdstream.Stream2D("flat_star_apcut",datadir_flat,anadir)
flat_star_apcut.fitsid=fitsid_flat_star.copy()
# aperture extraction
flat_star_apcut.band=band
print(flat_star_apcut.band,' band')
if band=='h' and flat_star_apcut.fitsid[0]%2==0:
flat_star_apcut.fitsid_increment()
ign_ord = [2, 15]
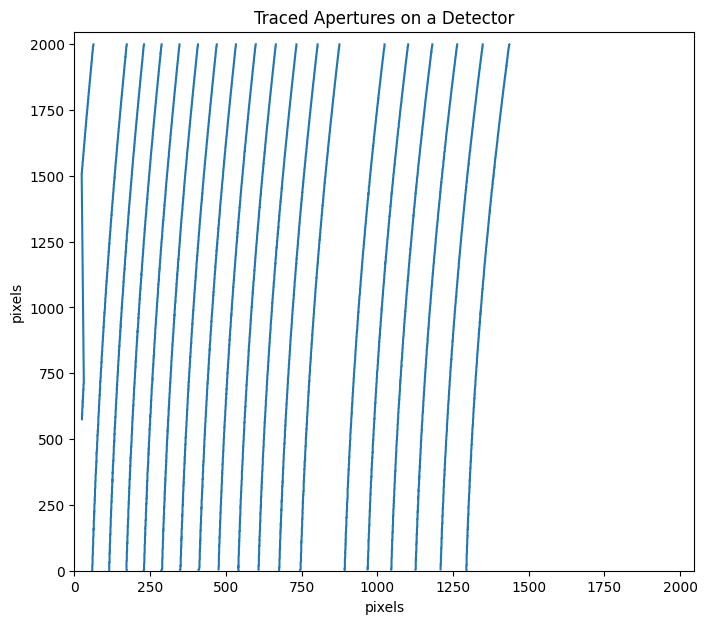
trace_mmf_apcut=flat_star_apcut.aptrace(cutrow = 1500,nap=19,ign_ord=ign_ord)
elif band=='y':
ign_ord = [51]
trace_mmf_apcut=flat_star_apcut.aptrace(cutrow = 1000,nap=50, ign_ord=ign_ord)
No fitsid yet.
h band
fitsid incremented: [41805, 41807, 41809, 41811, 41813, 41815, 41817, 41819, 41821, 41823, 41825, 41827, 41829, 41831, 41833, 41835, 41837, 41839, 41841, 41843, 41845, 41847, 41849, 41851, 41853, 41855, 41857, 41859, 41861, 41863, 41865, 41867, 41869, 41871, 41873, 41875, 41877, 41879, 41881, 41883, 41885, 41887, 41889, 41891, 41893, 41895, 41897, 41899, 41901, 41903]
median combine:
100%|█████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 50/50 [00:00<00:00, 1111.31it/s]
/Users/yuikasagi/git/pyird/src/pyird/image/aptrace.py:253: UserWarning: nap is not default value. default: nap = 42 for H / 102 for YJ.
warnings.warn(
cross-section: row 1502
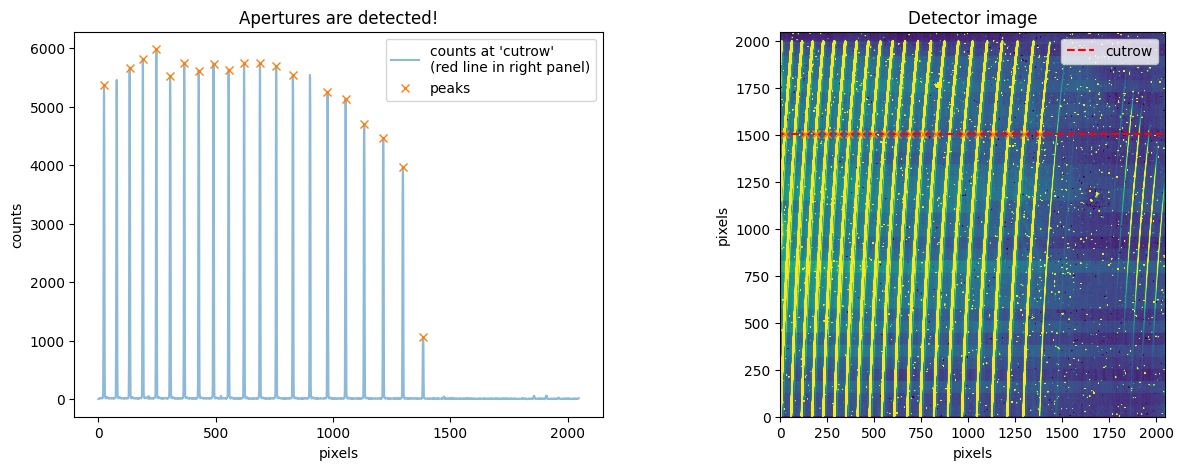
100%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 19/19 [00:03<00:00, 6.32it/s]
Additionally, you must set channelfile_path when performing
wavelength_calibration.
For example, (1) if you want to ignore orders 2 and 15, create a new
channel file in the same format as channel_H.list.
import importlib
import pandas as pd
# load the prepared file
channelfile_path_original = importlib.resources.files('pyird').joinpath('data/channel_H.list')
df_pixwavmap_original = pd.read_csv(channelfile_path_original)
df_pixwavmap_original
| ORDER | CHANNEL | WAVELENGTH | ST | |
|---|---|---|---|---|
| 0 | 1 | 905 | 1417.25 | 0 |
| 1 | 1 | 970 | 1417.86 | 0 |
| 2 | 1 | 1981 | 1425.31 | 0 |
| 3 | 2 | 370 | 1425.31 | 0 |
| 4 | 2 | 437 | 1426.08 | 0 |
| ... | ... | ... | ... | ... |
| 63 | 20 | 1319 | 1738.53 | 0 |
| 64 | 20 | 1333 | 1738.67 | 0 |
| 65 | 20 | 1555 | 1740.67 | 0 |
| 66 | 21 | 164 | 1744.97 | 0 |
| 67 | 21 | 1289 | 1758.93 | 0 |
68 rows × 4 columns
# Get indices to be dropped
mask_ind = df_pixwavmap_original[df_pixwavmap_original["ORDER"].isin(ign_ord)].index
# Drop the specified rows
df_pixwavmap_new = df_pixwavmap_original.drop(index=mask_ind)
df_pixwavmap_new
| ORDER | CHANNEL | WAVELENGTH | ST | |
|---|---|---|---|---|
| 0 | 1 | 905 | 1417.25 | 0 |
| 1 | 1 | 970 | 1417.86 | 0 |
| 2 | 1 | 1981 | 1425.31 | 0 |
| 6 | 3 | 689 | 1442.85 | 0 |
| 7 | 3 | 960 | 1445.57 | 1 |
| ... | ... | ... | ... | ... |
| 63 | 20 | 1319 | 1738.53 | 0 |
| 64 | 20 | 1333 | 1738.67 | 0 |
| 65 | 20 | 1555 | 1740.67 | 0 |
| 66 | 21 | 164 | 1744.97 | 0 |
| 67 | 21 | 1289 | 1758.93 | 0 |
63 rows × 4 columns
Save
df_pixwavmap_newto somewhere.
channelfile_path_new = "/path/to/save/new_channelfile.list"
df_pixwavmap_new.to_csv(channelfile_path_new, index=False)
Do not forget to set
channelfle_path.
thar.calibrate_wavelength(channelfile_path=channelfile_path_new, ign_ord=ign_ord)