🐱 Bayes Inference of a Real Spectrum of Luhman 16A (Direct LPF)¶
Update: Nov 10/2021, Hajime Kawahara
In this tutorial, we provide a tutorial for the HMC-NUTS fitting using NumPyro to the high-dispersion spectrum of Luhman 16A (Crossfield+2014) using the LPF direct. See also fidtbd_modit for the use of a rapid opacity calculator, MODIT. As the goal of this tutorial, we want to fit the exojax model to the high-dispersion data as
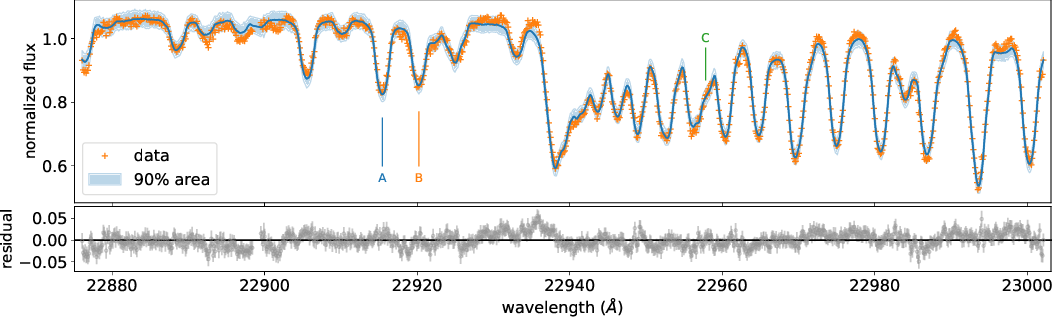
and get a posterior sampling. See the directories of examples/LUH16A/FidEMbu (Case I, w/o a GP) or examples/LUH16A/FidEMbug (Case II, w/ a GP) for the full python codes.
First, we import basic modules and some modules from jax,
# Basic modules
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# JAX
import jax.numpy as jnp
from jax import random
and, many modules and functions from ExoJAX.
# ExoJAX
from exojax.spec import initspec, planck, moldb, contdb, response, molinfo
from exojax.spec.lpf import xsvector, xsmatrix, exomol
from exojax.spec.exomol import gamma_exomol
from exojax.spec.hitrancia import read_cia, logacia
from exojax.spec.rtransfer import rtrun, dtauM, dtauCIA, nugrid, pressure_layer
from exojax.plot.atmplot import plot_maxpoint
from exojax.spec.evalline import reduceline_exomol
from exojax.spec.limb_darkening import ld_kipping
from exojax.utils.afunc import getjov_gravity
from exojax.utils.instfunc import R2STD
from exojax.utils.constants import RJ, pc
If you want to model a correlated noise by a gaussian process (GP), also call this one.
# CASE II
from exojax.utils.gpkernel import gpkernel_RBF
To fit the model to real high-resolution spectra, we usually need some information on absolute flux. Here, we just use the value from the mid-resolution spectrum.
# FLUX reference
Fabs_REF2=2.7e-12 #absolute flux (i.e. flux@10pc) erg/s/cm2/um Burgasser+ 1303.7283 @2.29um
fac0=RJ**2/((10.0*pc)**2) #nomralize by RJ
Fref=(2.29**2)*Fabs_REF2/fac0/1.e4 #erg/cm2/s/cm-1 @ 2.3um
Loading the real data of Luhman-16A by Crossfield+2014.
# Loading spectrum
dat=pd.read_csv("../data/luhman16a_spectra_detector1.csv",delimiter=",")
wavd=(dat["wavelength_micron"].values)*1.e4 #AA
nusd=1.e8/wavd[::-1]
fobs=(dat["normalized_flux"].values)[::-1]
err=(dat["err_normalized_flux"].values)[::-1]
Here we define the atmospheric layer (100 layers) and some qunatities for the atmospheric model.
# ATMOSPHERIC LAYER
Pref=1.0 # Reference pressure for a T-P model (bar)
NP=100
Parr, dParr, k=pressure_layer(NP=NP)
mmw=2.33 #mean molecular weight
ONEARR=np.ones_like(Parr) #ones_array for MMR
molmassCO=molinfo.molmass("CO") #molecular mass (CO)
molmassH2O=molinfo.molmass("H2O") #molecular mass (H2O)
Assuming the instrumental resolution… Yes, beta is the standard deviation of the Gaussian.
# Instrument
beta=R2STD(100000.) #std of gaussian from R=100000.
Here, we set the wavenumber grid, with the target range between ws and we AA, but having a margin +- 5 AA.
# Loading Molecular datanase and Reducing Molecular Lines
Nx=4500 # number of wavenumber bins (nugrid) for fit
ws=22876.0 # AA
we=23010.0 # AA
nus,wav,res=nugrid(ws-5.0,we+5.0,Nx,unit="AA")
Some masking.
# Masking data
mask=(ws<wavd[::-1])*(wavd[::-1]<we) # data fitting range
mask=mask*((22898.5>wavd[::-1])+(wavd[::-1]>22899.5)) # Additional mask to remove a strong telluric
fobsx=fobs[mask]
nusdx=nusd[mask]
wavdx=1.e8/nusdx[::-1]
errx=err[mask]
Loading exomol databases for CO and H2O…
# Loading molecular database
mdbCO=moldb.MdbExomol('.database/CO/12C-16O/Li2015',nus)
mdbH2O=moldb.MdbExomol('.database/H2O/1H2-16O/POKAZATEL',nus,crit=1.e-46)
and CIA from HITRAN.
# LOADING CIA
mmrH2=0.74
mmrHe=0.25
molmassH2=molinfo.molmass("H2")
molmassHe=molinfo.molmass("He")
vmrH2=(mmrH2*mmw/molmassH2)
vmrHe=(mmrHe*mmw/molmassHe)
cdbH2H2=contdb.CdbCIA('.database/H2-H2_2011.cia',nus)
cdbH2He=contdb.CdbCIA('.database/H2-He_2011.cia',nus)
This example uses the direct LPF. So, one might reduce weak lines to save the computational time. But how? we have a simple function for that purpose. Assuming a constant T model, we can exclude the lines below the CIA photosphere.
# Reducing Molecular Lines
def Tmodel(Parr,T0):
""" Constant T model
"""
return T0*np.ones_like(Parr)
# Reference physical quantities
g=10**(5.0)
maxMMR_CO=0.01
maxMMR_H2O=0.005
# CO
mask_CO,maxcf,maxcia=reduceline_exomol(mdbCO,Parr,dParr,mmw,g,vmrH2,cdbH2H2,maxMMR_CO,molmassCO,Tmodel,[1700.0]) #only 1700K
plot_maxpoint(mask_CO,Parr,maxcf,maxcia,mol="CO")
plt.savefig("maxpoint_CO.pdf", bbox_inches="tight", pad_inches=0.0)
# H2O
T0xarr=list(range(500,1800,100))
mask_H2O,maxcf,maxcia=reduceline_exomol(mdbH2O,Parr,dParr,mmw,g,vmrH2,cdbH2H2,maxMMR_H2O,molmassH2O,Tmodel,T0xarr) #only 1700K
plot_maxpoint(mask_H2O,Parr,maxcf,maxcia,mol="H2O")
plt.savefig("maxpoint_H2O.pdf", bbox_inches="tight", pad_inches=0.0)
The initialization of the direct LPF (or just precompute nu matrix).
# Initialization of direct LPF
numatrix_CO=initspec.init_lpf(mdbCO.nu_lines,nus)
numatrix_H2O=initspec.init_lpf(mdbH2O.nu_lines,nus)
We are now ready for an HMC-NUTS fitting!
# HMC-NUTS FITTING PART
from numpyro import sample
import numpyro.distributions as dist
from numpyro.infer import MCMC, NUTS
from numpyro.infer import Predictive
from numpyro.diagnostics import hpdi
# Some constants for fitting
baseline=1.07 #(baseline for a CIA photosphere in the observed (normaized) spectrum)
maxMMR_CO=0.01
maxMMR_H2O=0.005
Define the model.
# Model
def model_c(nu1,y1,e1):
Rp = sample('Rp', dist.Uniform(0.5,1.5))
Mp = sample('Mp', dist.Normal(33.5,0.3))
RV = sample('RV', dist.Uniform(26.0,30.0))
MMR_CO = sample('MMR_CO', dist.Uniform(0.0,maxMMR_CO))
MMR_H2O = sample('MMR_H2O', dist.Uniform(0.0,maxMMR_H2O))
T0 = sample('T0', dist.Uniform(1000.0,1700.0))
alpha = sample('alpha', dist.Uniform(0.05,0.15))
vsini = sample('vsini', dist.Uniform(10.0,20.0))
# Kipping Limb Darkening Prior
q1 = sample('q1', dist.Uniform(0.0,1.0))
q2 = sample('q2', dist.Uniform(0.0,1.0))
u1,u2=ld_kipping(q1,q2)
If you want to model a correlated noise by a GP, set the GP hyperparameters
#def model_c(nu1,y1,e1): (continued)
# CASE II
#GP
logtau = sample('logtau', dist.Uniform(-1.5,0.5)) #tau=1 <=> 5A
tau=10**(logtau)
loga = sample('loga', dist.Uniform(-4.0,-2.0))
a=10**(loga)
Otherwise, define sigma in an independent gaussian:
#def model_c(nu1,y1,e1): (continued)
# CASE I
sigma = numpyro.sample('sigma', dist.Exponential(10.0))
Set gravity using radius and mass in the Jovian unit.
#def model_c(nu1,y1,e1): (continued)
#gravity
g=getjov_gravity(Rp,Mp)
And here we assume a power-law type temperature model. This can be relaxed.
#def model_c(nu1,y1,e1): (continued)
#T-P model//
Tarr = T0*(Parr/Pref)**alpha
spec.lpf.exomol is a convenient way to obtain the quantities for line profile.
#def model_c(nu1,y1,e1): (continued)
#CO
SijM_CO,gammaLM_CO,sigmaDM_CO=exomol(mdbCO,Tarr,Parr,molmassCO)
xsm_CO=xsmatrix(numatrix_CO,sigmaDM_CO,gammaLM_CO,SijM_CO)
dtaumCO=dtauM(dParr,xsm_CO,MMR_CO*ONEARR,molmassCO,g)
#H2O
SijM_H2O,gammaLM_H2O,sigmaDM_H2O=exomol(mdbH2O,Tarr,Parr,molmassH2O)
xsm_H2O=xsmatrix(numatrix_H2O,sigmaDM_H2O,gammaLM_H2O,SijM_H2O)
dtaumH2O=dtauM(dParr,xsm_H2O,MMR_H2O*ONEARR,molmassH2O,g)
#CIA
dtaucH2H2=dtauCIA(nus,Tarr,Parr,dParr,vmrH2,vmrH2,\
mmw,g,cdbH2H2.nucia,cdbH2H2.tcia,cdbH2H2.logac)
dtaucH2He=dtauCIA(nus,Tarr,Parr,dParr,vmrH2,vmrHe,\
mmw,g,cdbH2He.nucia,cdbH2He.tcia,cdbH2He.logac)
dtau=dtaumCO+dtaumH2O+dtaucH2H2+dtaucH2He
sourcef = planck.piBarr(Tarr,nus)
Ftoa=Fref/Rp**2
F0=rtrun(dtau,sourcef)/baseline/Ftoa
Frot=response.rigidrot(nus,F0,vsini,u1,u2)
mu=response.ipgauss_sampling(nu1,nus,Frot,beta,RV)
Here, in the case of a GP modeling of the noise, just define the GP kernel and use dist.MultivariateNormal. So simple!
#def model_c(nu1,y1,e1): (continued)
# CASE II
cov=gpkernel_RBF(nu1,tau,a,e1)
sample("y1", dist.MultivariateNormal(loc=mu, covariance_matrix=cov), obs=y1)
Or you prefer an independent Gaussan?
#def model_c(nu1,y1,e1): (continued)
# CASE I
cov=gpkernel_RBF(nu1,tau,a,e1)
errall=jnp.sqrt(e1**2+sigma**2)
sample(tag, dist.Normal(mu, errall), obs=y)
Then, run the HMC-NUTS. Note that we here use forward mode (forward differentiation) by ‘forward_mode_differentiation=True’ in NUTS. Since ExoJAX v1.1, we can also use the reverse mode ‘forward_mode_differentiation=False’ in NUTS.
#Running a HMC-NUTS
rng_key = random.PRNGKey(0)
rng_key, rng_key_ = random.split(rng_key)
num_warmup, num_samples = 500, 1000
kernel = NUTS(model_c,forward_mode_differentiation=True)
mcmc = MCMC(kernel, num_warmup=num_warmup, num_samples=num_samples)
mcmc.run(rng_key_, nu1=nusdx, y1=fobsx, e1=errx)
print("end HMC")
That’s all! The rest part is just for saving and plotting.
# Post-processing
posterior_sample = mcmc.get_samples()
np.savez("npz/savepos.npz",[posterior_sample])
pred = Predictive(model_c,posterior_sample,return_sites=["y1"])
nu = nus
predictions = pred(rng_key_,nu1=nu,y1=None,e1=errx)
median_mu = jnp.median(predictions["y1"],axis=0)
hpdi_mu = hpdi(predictions["y1"], 0.9)
np.savez("npz/saveplotpred.npz",[wavdx,fobsx,errx,median_mu,hpdi_mu])
Arviz is very useful for plotting, such as the corner plot, the trace plot and so on.
# ARVIZ part
import arviz
rc = {
"plot.max_subplots": 1024,
}
try:
arviz.rcParams.update(rc)
arviz.plot_pair(arviz.from_numpyro(mcmc),kind='kde',divergences=False,marginals=True)
plt.savefig("npz/cornerall.png")
except:
print("failed corner")
try:
pararr=["Mp","Rp","T0","alpha","MMR_CO","MMR_H2O","vsini","RV","q1","q2","logtau","loga"]
arviz.plot_trace(mcmc, var_names=pararr)
plt.savefig("npz/trace.png")
except:
print("failed trace")
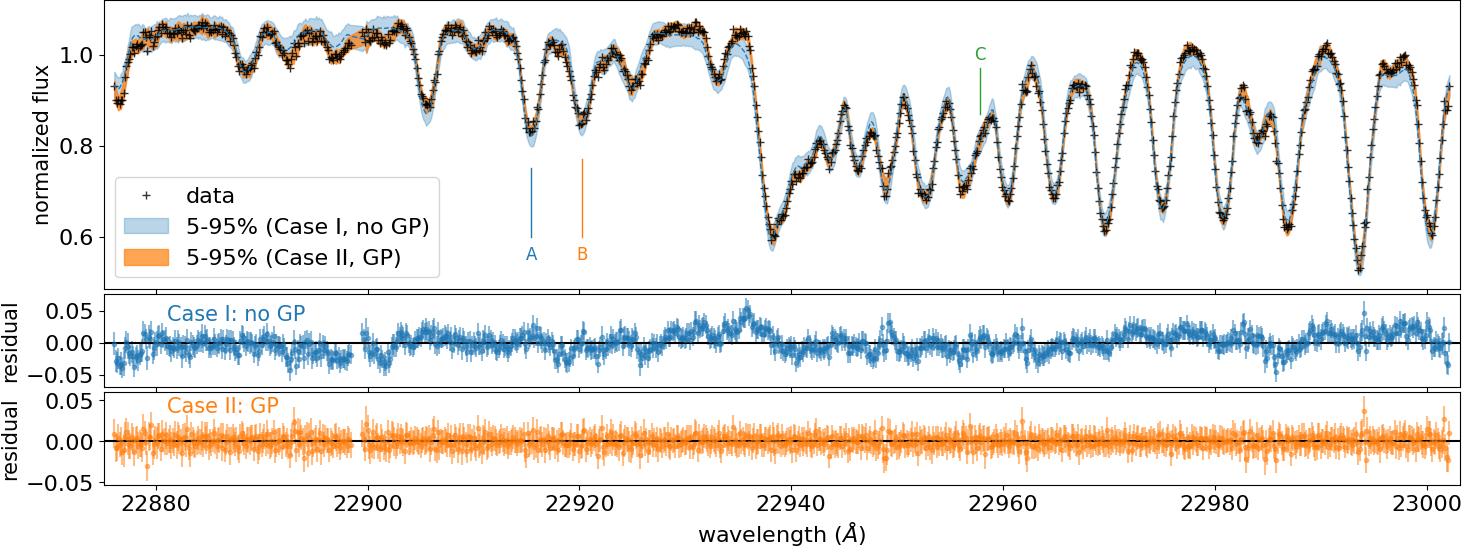
Credible interval for the GP case (CASE II)¶
Here, we show an example to compute the credible interval of the prdiction including a GP. See Appendix F in Paper I (Kawahara+2021) for more details.
The probability of the prediction \({\bf d}^\ast\) for an arbitrary wavenumber vector \({\bf \nu}^\ast\) conditioned on the given data \({\bf d}\) is expressed as
\(p({\bf d}^\ast|{\bf d}) = {\mathcal N} ({F}({\bf \nu}^\ast) + K_{\times}^\top K_\sigma^{-1} ({\bf d} - {F}({\bf \nu})) K_{\ast,\sigma} - K_\times^\top K_\sigma^{-1} K_\times)\)
where
\((K_{\times})_{ij} = a \, {k}(|\nu_i-\nu^\ast_j|;\tau)\)
\((K_{\sigma})_{ij} = a \, {k}(|\nu_i-\nu_j|;\tau) + \sigma_{e,i}^2 \delta_{ij}\)
\((K_{\ast,\sigma})_{ij} = a \, {k}(|\nu^\ast_i-\nu^\ast_j|;\tau) + (\sigma_{e,i}^\ast)^2 \delta_{ij}\)
where \(\delta_{ij}\) is the Kronecker delta. Then the code should be like the below.
mu = #the spectral model (skipeed here)
cov = gpkernel_RBF(t,t,tau,a) + jnp.diag(err**2)
covx= gpkernel_RBF(t,td,tau,a)
covxx = gpkernel_RBF(td,td,tau,a) + jnp.diag(err**2)
A=scipy.linalg.solve(cov,fobsx-mu,assume_a="pos")
IKw = scipy.linalg.inv(cov)
gp=covx@A
newcov=covxx - covx@IKw@covx.T
For instance, the same wavenumber grid for t and td can be used.
t=nusdx
td=nusdx
An HMC simulation provides a sampling of the other parameters \({\bf \theta}^\dagger\) than the GP parameters. Then, the prediction can be sampled by
\({\bf d}^\ast_k \sim {\mathcal N} ({F}({\bf \nu}^\ast; {\bf \theta}^\dagger_k) + K_{\times}^\top K_\sigma^{-1} ({\bf d} - {F}({\bf \nu}; {\bf \theta}^\dagger_k)) K_{\ast,\sigma} - K_\times^\top K_\sigma^{-1} K_\times)\)
where
\({\bf \theta}^\dagger_k\) is the k-th sampling of \({\bf \theta}^\dagger\) . The credible interval can be computed using the sampling given by the anove Equation. This prediction includes independent Gaussian noise \(\sigma_{e,i}^\ast\) . When we adopt \(\sigma_{e,i}^\ast = 0\) , This equation simply provides the prediction of the spectral model + trend.
Then, one can get a sampling using multivariate_normal in scipy.stat as
from scipy.stats import multivariate_normal as smn
mkgp = smn(mean=gp ,cov=newcov , allow_singular =True).rvs(1).T
Computing the HPDI, we get the orange area in the following figure:
Layer-by-Layer Modelling¶
We can also model the temperature profile (and a VMR) by a layer-by-layer model instead of the power-law model. In this case, add a small identity matrix to the RBF kernel to prevent overflow such as
def modelcov(t,tau,a):
fac=1.e-5 #small value
Dt = t - jnp.array([t]).T
K=a*jnp.exp(-(Dt)**2/2/(tau**2))+a*fac*jnp.identity(NP)
return K
Then, use dist.MultivariateNormal to model the prior of the temperature layers:
# Model
def model_c(nu1,y1,e1):
Rp = sample('Rp', dist.Uniform(0.5,1.5))
Mp = sample('Mp', dist.Normal(33.5,0.3))
RV = sample('RV', dist.Uniform(26.0,30.0))
MMR_CO = sample('MMR_CO', dist.Uniform(0.0,maxMMR_CO))
MMR_H2O = sample('MMR_H2O', dist.Uniform(0.0,maxMMR_H2O))
#Layer-by-layer T-P model
lnsT=6.0
sT=10**lnsT
lntaup=0.5
taup=10**lntaup
cov=modelcov(lnParr,taup,sT)
T0 = numpyro.sample('T0', dist.Uniform(1000,1600))
Tarr=numpyro.sample("Tarr", dist.MultivariateNormal(loc=ONEARR, covariance_matrix=cov))+T0
#(continued)