CKD Transmission Tutorial (load only): ArtTransPure with OpaCKD
Hajime Kawahara with Claude Code, September 24th (2025)
This tutorial demonstrates how to use the Correlated K-Distribution (CKD) method for atmospheric transmission calculations with ExoJAX, by loading exisiting saved data. We also run a simple HMC-NUTS using generated data.
# Import required packages
import numpy as np
import matplotlib.pyplot as plt
from jax import config
# ExoJAX imports
from exojax.test.emulate_mdb import mock_wavenumber_grid
from exojax.opacity import OpaCKD
from exojax.rt import ArtTransPure
# Enable 64-bit precision for accurate calculations
config.update("jax_enable_x64", True)
print("ExoJAX CKD Tutorial: Transmission Spectroscopy")
print("=============================================")
ExoJAX CKD Tutorial: Transmission Spectroscopy
=============================================
1. Setup Atmospheric Model and Molecular Database
First, we’ll set up our atmospheric model for transmission spectroscopy calculations.
# Setup wavenumber grid and molecular database
nu_grid, wav, res = mock_wavenumber_grid()
print(f"Wavenumber grid: {len(nu_grid)} points from {nu_grid[0]:.1f} to {nu_grid[-1]:.1f} cm⁻¹")
print(f"Spectral resolution: {res:.1f}")
# Setup atmospheric radiative transfer for transmission
art = ArtTransPure(
pressure_top=1.0e-8,
pressure_btm=1.0e2,
nlayer=50, # Fewer layers for transmission calculations
integration="simpson" # Simpson integration for better accuracy
)
print(f"Atmospheric layers: {art.nlayer}")
print(f"Pressure range: {art.pressure_top:.1e} - {art.pressure_btm:.1e} bar")
print(f"Integration method: {art.integration}")
xsmode = modit xsmode assumes ESLOG in wavenumber space: xsmode=modit Your wavelength grid is in * ascending * order The wavenumber grid is in ascending order by definition. Please be careful when you use the wavelength grid. Wavenumber grid: 20000 points from 4329.0 to 4363.0 cm⁻¹ Spectral resolution: 2556525.8 integration: simpson Simpson integration, uses the chord optical depth at the lower boundary and midppoint of the layers. Atmospheric layers: 50 Pressure range: 1.0e-08 - 1.0e+02 bar Integration method: simpson
/home/kawahara/exojax/src/exojax/utils/grids.py:85: UserWarning: Both input wavelength and output wavenumber are in ascending order.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/grids.py:85: UserWarning: Both input wavelength and output wavenumber are in ascending order.
warnings.warn(
/home/kawahara/exojax/src/exojax/rt/common.py:40: UserWarning: nu_grid is not given. specify nu_grid when using 'run'
warnings.warn(
2. Define Atmospheric and Planetary Parameters
We’ll create atmospheric profiles and define planetary parameters for transmission calculations.
# Create atmospheric profiles
Tarr = np.linspace(1000.0, 1500.0, 50) # Temperature profile
mmr_arr = np.full(50, 0.1) # Constant H2O mixing ratio
mean_molecular_weight = np.full(50, 2.33) # Mean molecular weight (H2-dominated)
# Planetary parameters (Jupiter-like)
radius_btm = 6.9e9 # Planet radius at bottom of atmosphere (cm)
gravity = 2478.57 # Surface gravity (cm/s²)
# Plot atmospheric profiles
fig, (ax1, ax2, ax3) = plt.subplots(1, 3, figsize=(15, 5))
# Temperature profile
ax1.semilogy(Tarr, art.pressure)
ax1.set_xlabel('Temperature (K)')
ax1.set_ylabel('Pressure (bar)')
ax1.set_title('Temperature Profile')
ax1.grid(True, alpha=0.3)
ax1.invert_yaxis()
# Mixing ratio profile
ax2.semilogy(mmr_arr, art.pressure)
ax2.set_xlabel('H₂O Mixing Ratio')
ax2.set_ylabel('Pressure (bar)')
ax2.set_title('H₂O Mixing Ratio Profile')
ax2.grid(True, alpha=0.3)
ax2.invert_yaxis()
# Mean molecular weight profile
ax3.semilogy(mean_molecular_weight, art.pressure)
ax3.set_xlabel('Mean Molecular Weight (amu)')
ax3.set_ylabel('Pressure (bar)')
ax3.set_title('Mean Molecular Weight Profile')
ax3.grid(True, alpha=0.3)
ax3.invert_yaxis()
plt.tight_layout()
plt.show()
print(f"Temperature range: {np.min(Tarr):.0f} - {np.max(Tarr):.0f} K")
print(f"H2O mixing ratio: {mmr_arr[0]:.1f} (constant)")
print(f"Mean molecular weight: {mean_molecular_weight[0]:.2f} amu (constant)")
print(f"Planet radius: {radius_btm/6.9e9:.1f} R_Jupiter")
print(f"Surface gravity: {gravity:.0f} cm/s² ({gravity/2478.57:.1f} × Jupiter)")
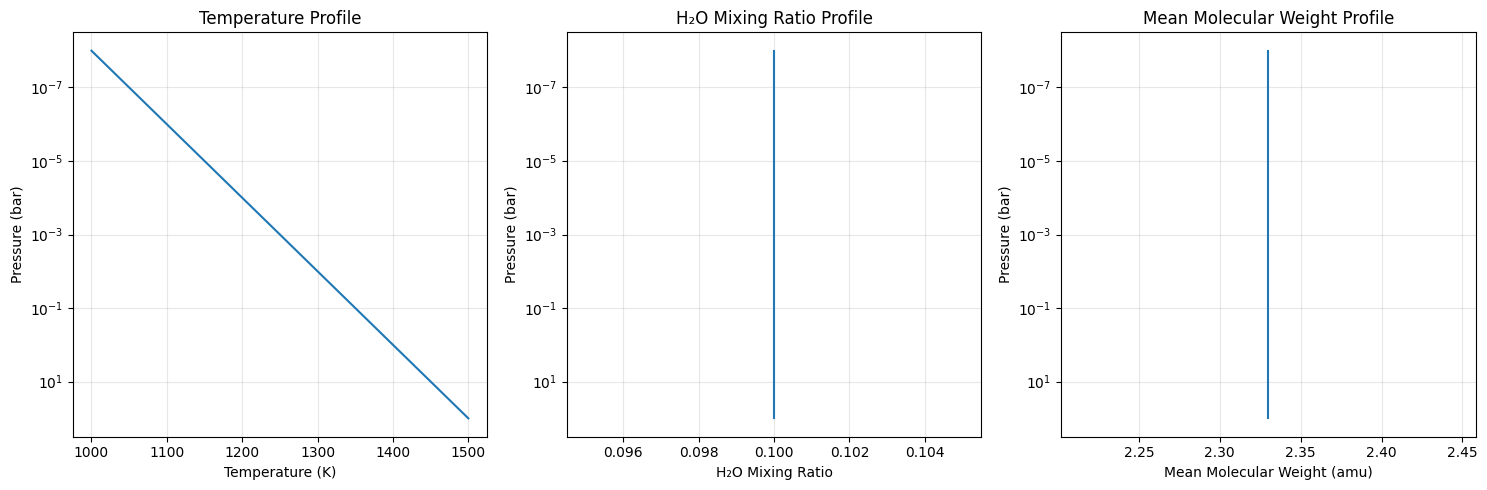
Temperature range: 1000 - 1500 K
H2O mixing ratio: 0.1 (constant)
Mean molecular weight: 2.33 amu (constant)
Planet radius: 1.0 R_Jupiter
Surface gravity: 2479 cm/s² (1.0 × Jupiter)
3. Setup CKD Opacity Calculator and Compute Transmission using the Saved Table Data
Now we’ll directly load the CKD opacity table data and compute the CKD transmission spectrum.
opa_ckd = OpaCKD.from_saved_tables("ckd_h2o.npz") #one liner, no initialization needed
# Alternatively, load only the CKD object and then load tables
#ckd = OpaCKD.load_only()
#ckd.load_tables("ckd_h2o.npz")
molmass = 18.02 # Molecular mass of H2O (g/mol)
print(f"CKD Opacity Calculator Setup:")
print(f" Number of g-ordinates (Ng): {opa_ckd.Ng}")
print(f" Band width: {opa_ckd.band_width}")
print(f" Number of spectral bands: {len(opa_ckd.nu_bands)}")
print(f" Spectral range: {opa_ckd.nu_bands[0]:.1f} - {opa_ckd.nu_bands[-1]:.1f} cm⁻¹")
# Pre-compute CKD tables on temperature-pressure grid
print("\nPre-computing CKD tables...")
T_grid = np.linspace(np.min(Tarr), np.max(Tarr), 10)
P_grid = np.logspace(np.log10(np.min(art.pressure)), np.log10(np.max(art.pressure)), 10)
# Get CKD cross-section tensor and compute CKD spectrum
print("Computing CKD transmission spectrum...")
xs_ckd = opa_ckd.xstensor_ckd(Tarr, art.pressure)
dtau_ckd = art.opacity_profile_xs_ckd(xs_ckd, mmr_arr, molmass, gravity)
transit_ckd = art.run_ckd(dtau_ckd, Tarr, mean_molecular_weight, radius_btm, gravity, opa_ckd.ckd_info.weights)
print(f"CKD spectrum computed!")
print(f"CKD transit range: [{np.min(transit_ckd):.6f}, {np.max(transit_ckd):.6f}]")
CKD Opacity Calculator Setup:
Number of g-ordinates (Ng): 16
Band width: 0.5
Number of spectral bands: 68
Spectral range: 4329.3 - 4362.8 cm⁻¹
Pre-computing CKD tables...
Computing CKD transmission spectrum...
CKD spectrum computed!
CKD transit range: [1.042467, 1.071651]
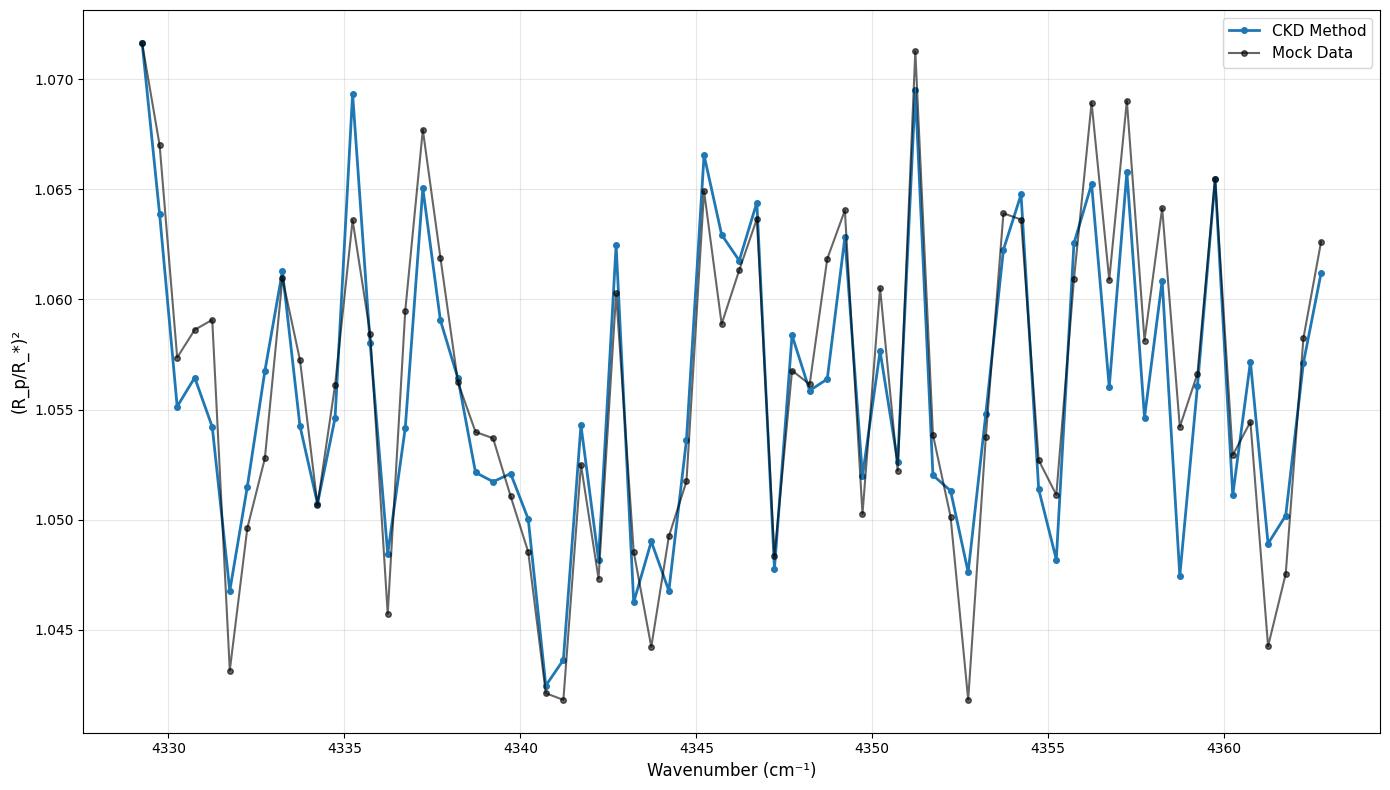
4. Generate Mock Data
3#make mock data
from numpy.random import default_rng
rng = default_rng(seed=12)
sigma = 0.003
mock_data = transit_ckd + rng.normal(0, sigma, len(transit_ckd))
# Create comparison plot
plt.figure(figsize=(14, 8))
plt.plot(opa_ckd.nu_bands, transit_ckd,
'o-', label="CKD Method",
markersize=4, linewidth=2, color='C0')
plt.plot(opa_ckd.nu_bands, mock_data,
'o-', label="Mock Data",
markersize=4, color='black', alpha=0.6)
plt.xlabel('Wavenumber (cm⁻¹)', fontsize=12)
plt.ylabel('(R_p/R_*)²', fontsize=12)
plt.legend(fontsize=11)
plt.grid(True, alpha=0.3)
plt.tight_layout()
plt.show()
5. Runs HMC-NUTS!
import jax.numpy as jnp
def fspec(mmr_const):
mmr_arr = jnp.full(50, mmr_const) # Constant H2O mixing ratio
xs_ckd = opa_ckd.xstensor_ckd(Tarr, art.pressure)
dtau_ckd = art.opacity_profile_xs_ckd(xs_ckd, mmr_arr, molmass, gravity)
mu = art.run_ckd(dtau_ckd, Tarr, mean_molecular_weight, radius_btm, gravity, opa_ckd.ckd_info.weights)
return mu
plt.plot(opa_ckd.nu_bands, fspec(0.1), 'o-', label="CKD Method (mmr=0.1)", markersize=4, linewidth=2, color='C0')
plt.plot(opa_ckd.nu_bands, fspec(0.01), 'o-', label="CKD Method (mmr=0.05)", markersize=4, linewidth=2, color='C1')
[<matplotlib.lines.Line2D at 0x7799b0194ee0>]
from numpyro.infer import MCMC, NUTS
import numpyro.distributions as dist
import numpyro
from jax import random
def model_prob(spectrum):
#atmospheric/spectral model parameters priors
mmr = numpyro.sample('MMR', dist.Uniform(0.0, 0.3))
mu = fspec(mmr)
#noise model parameters priors
sigmain = numpyro.sample('sigmain', dist.Exponential(1.e0))
numpyro.sample('spectrum', dist.Normal(mu, sigmain), obs=spectrum)
rng_key = random.PRNGKey(0)
rng_key, rng_key_ = random.split(rng_key)
num_warmup, num_samples = 500, 1000
#kernel = NUTS(model_prob, forward_mode_differentiation=True)
kernel = NUTS(model_prob, forward_mode_differentiation=False)
mcmc = MCMC(kernel, num_warmup=num_warmup, num_samples=num_samples)
mcmc.run(rng_key_, spectrum=mock_data)
mcmc.print_summary()
sample: 100%|██████████| 1500/1500 [00:10<00:00, 139.48it/s, 3 steps of size 8.29e-01. acc. prob=0.92]
mean std median 5.0% 95.0% n_eff r_hat
MMR 0.11 0.01 0.11 0.09 0.13 508.97 1.00
sigmain 0.00 0.00 0.00 0.00 0.00 1082.87 1.00
Number of divergences: 0
from numpyro.diagnostics import hpdi
from numpyro.infer import Predictive
import jax.numpy as jnp
# SAMPLING
posterior_sample = mcmc.get_samples()
pred = Predictive(model_prob, posterior_sample, return_sites=['spectrum'])
predictions = pred(rng_key_, spectrum=None)
median_mu1 = jnp.median(predictions['spectrum'], axis=0)
hpdi_mu1 = hpdi(predictions['spectrum'], 0.9)
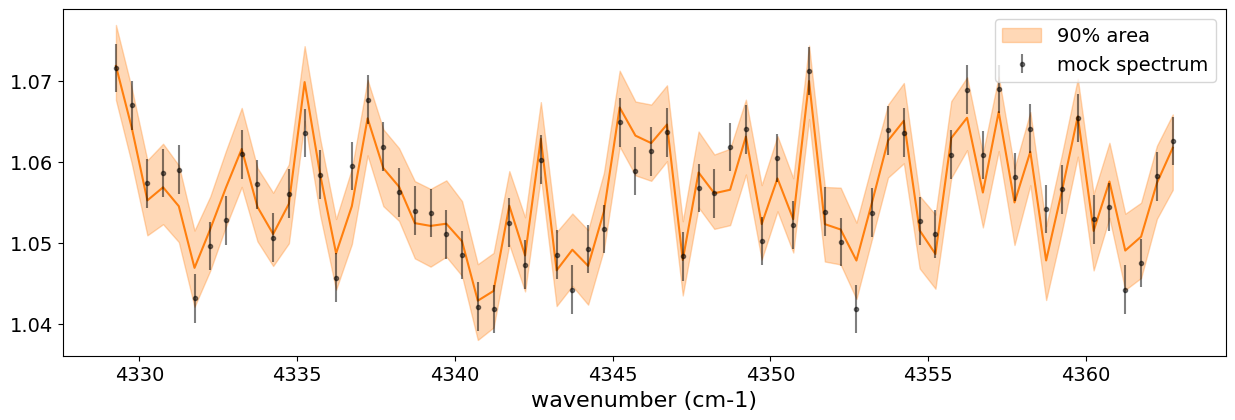
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(15, 4.5))
ax.plot(opa_ckd.nu_bands, median_mu1, color='C1')
ax.fill_between(opa_ckd.nu_bands,
hpdi_mu1[0],
hpdi_mu1[1],
alpha=0.3,
interpolate=True,
color='C1',
label='90% area')
ax.errorbar(opa_ckd.nu_bands, mock_data, sigma, fmt=".", label="mock spectrum", color="black",alpha=0.5)
plt.xlabel('wavenumber (cm-1)', fontsize=16)
plt.legend(fontsize=14)
plt.tick_params(labelsize=14)
plt.show()
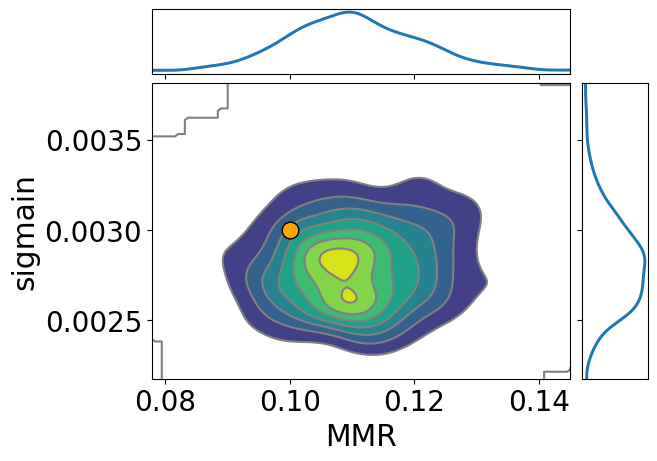
import arviz
pararr = ["MMR", "sigmain"]
arviz.plot_pair(
arviz.from_numpyro(mcmc),
kind="kde",
divergences=False,
marginals=True,
reference_values={
"MMR": 0.1,
"sigmain": 0.003,
},
reference_values_kwargs={
"marker": "o",
"markersize": 12,
"linestyle": "None",
"color": "orange",
},
textsize=20,
)
plt.show()