Reduces Device Memory Usage by Dividing and Stitching the Wavenumber Grid (\(\nu\) - stitching)
Hajime Kawahara September 25th (2025)
Here, we explain the method of reducing GPU device memory usage by
dividing the wavenumber grid (\(\nu\) - stitching). This approach is
particularly effective for transmission spectroscopy, where
opart cannot be used. When applying this
method, forward-mode
differentiation
should be used for derivatives. While \(\nu\) - stitching can be
performed manually, we describe the
procedure using \(\nu\) - stitching in OpaPremodit in this
section.
Currently, the only opacity calculator that supports
\(\nu\)-stitching is PreMODIT (v2.0), which can be used with
OpaPremodit.
from exojax.opacity import OpaPremodit
from exojax.utils.grids import wavenumber_grid
from exojax.database.exomol.api import MdbExomol
from exojax.rt import ArtTransPure
import jax.numpy as jnp
from jax import config
config.update("jax_enable_x64", True)
In this example, the OH molecule is used to compute opacity over the 1.7–2.3 micron range, divided into 300,000 segments, with 300 atmospheric layers. At the time of creating this notebook, the computation was performed on a gaming laptop with 8GB of device memory.
N=300000
nus, wav, res = wavenumber_grid(17000.0, 23000.0, N, unit="AA", xsmode="premodit")
print("resolution=",res)
mdb = MdbExomol(".database/OH/16O-1H/MoLLIST/", nus)
molmass = mdb.molmass
xsmode = premodit
xsmode assumes ESLOG in wavenumber space: xsmode=premodit
Your wavelength grid is in * descending * order
The wavenumber grid is in ascending order by definition.
Please be careful when you use the wavelength grid.
resolution= 992451.1535950146
HITRAN exact name= (16O)H
radis engine = vaex
Molecule: OH
Isotopologue: 16O-1H
ExoMol database: None
Local folder: .database/OH/16O-1H/MoLLIST
Transition files:
=> File 16O-1H__MoLLIST.trans
Broadener: H2
The default broadening parameters are used.
/home/kawahara/exojax/src/exojax/utils/grids.py:85: UserWarning: Both input wavelength and output wavenumber are in ascending order. warnings.warn( /home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname. warnings.warn( /home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname. warnings.warn( /home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname. warnings.warn( /home/kawahara/anaconda3/lib/python3.10/site-packages/radis/api/exomolapi.py:1626: UserWarning: Could not load 16O-1H__H2.broad. The default broadening parameters are used. warnings.warn(
art = ArtTransPure(pressure_top=1.e-10, pressure_btm=1.e1, nlayer=300) #300 OK
Tarr = jnp.ones_like(art.pressure)*3000.0
Parr = art.pressure
integration: simpson
Simpson integration, uses the chord optical depth at the lower boundary and midppoint of the layers.
/home/kawahara/exojax/src/exojax/rt/common.py:40: UserWarning: nu_grid is not given. specify nu_grid when using 'run'
warnings.warn(
Now, we proceed with the opacity calculation. Here, the wavenumber range
is divided into 20 segments, and the opacity is computed by summing over
them using OLA. The parameter cutwing specifies where to truncate
the line wings. In this case, cutwing is set to 0.015, meaning the
truncation occurs at 0.015 times the wavenumber grid spacing, which
corresponds to approximately 20 cm-1.
Please refer to this section for the mechanism of OLA-based combination.
ndiv=20
snap = mdb.to_snapshot()
del mdb
opas = OpaPremodit.from_snapshot(snap, nus, nstitch=ndiv, auto_trange=[500,1300], cutwing = 0.015)
xsm_s = opas.xsmatrix(Tarr, Parr)
default elower grid trange (degt) file version: 2
Robust range: 485.7803992045456 - 1334.4906506037173 K
max value of ngamma_ref_grid : 9.677379608844298
min value of ngamma_ref_grid : 7.156060542679381
ngamma_ref_grid grid : [7.15606022 7.91349981 8.75111089 9.67738056]
max value of n_Texp_grid : 0.5
min value of n_Texp_grid : 0.5
n_Texp_grid grid : [0.49999997 0.50000006]
uniqidx: 100%|██████████| 2/2 [00:00<00:00, 4807.23it/s]
Premodit: Twt= 1049.0651485510987 K Tref= 539.7840596059918 K
Making LSD:|####################| 100%
2025-09-25 08:26:58.752216: W external/xla/xla/hlo/transforms/simplifiers/hlo_rematerialization.cc:3023] Can't reduce memory use below 3.15GiB (3379151558 bytes) by rematerialization; only reduced to 3.52GiB (3775929320 bytes), down from 3.52GiB (3775948936 bytes) originally
You can check the wing-cut wavenumber \(\Delta \nu \sim 20\) cm-1.
from exojax.utils.astrofunc import gravity_jupiter
mmr = jnp.ones_like(Parr)*0.01
g = gravity_jupiter(1.0,1.0)
dtau = art.opacity_profile_xs(xsm_s,mmr,molmass,g)
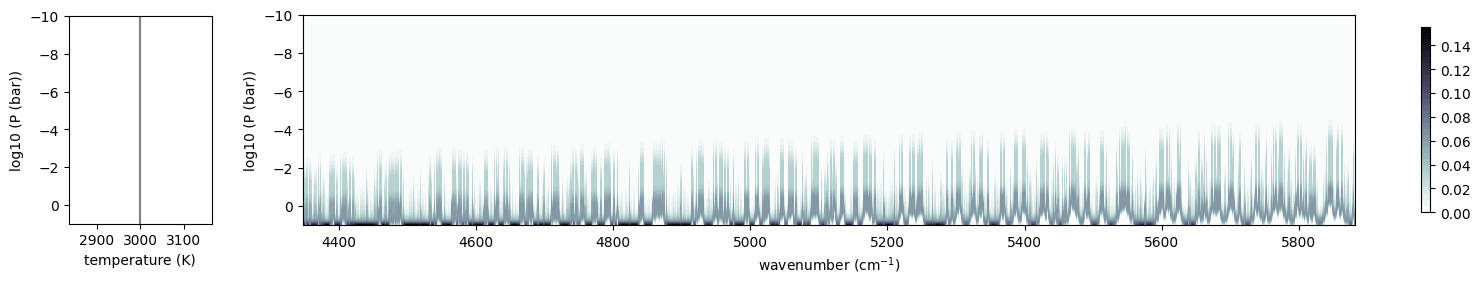
Let’s check the contribution function. It is clear that lines are present across a wide wavenumber range.
from exojax.plot.atmplot import plotcf
cf = plotcf(nus, dtau, Tarr, Parr, art.dParr)
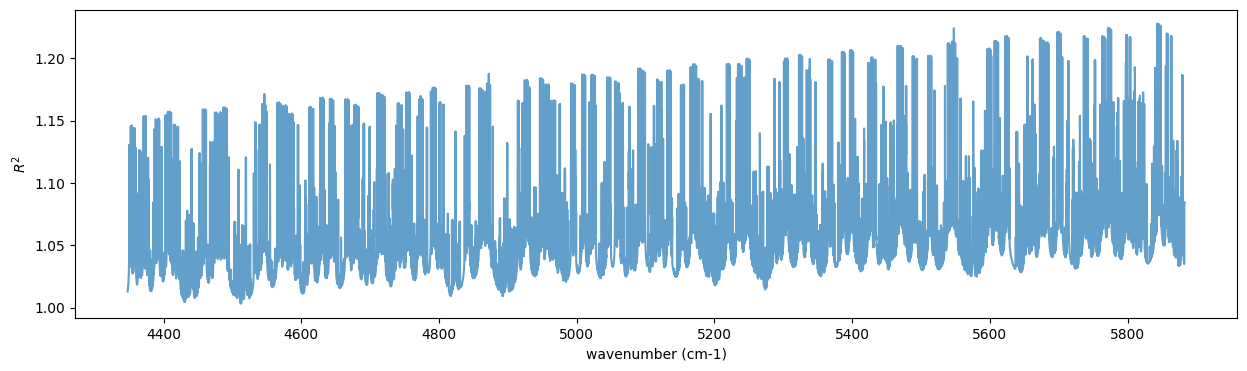
Let’s calculate the transmitted light spectrum.
from exojax.utils.constants import RJ
mmw = jnp.ones_like(Parr)*2.0
r2 = art.run(dtau, Tarr, mmw, RJ, g)
import matplotlib.pyplot as plt
plt.figure(figsize=(15, 4))
plt.plot(nus, r2, alpha=0.7)
plt.ylabel("$R^2$")
plt.xlabel("wavenumber (cm-1)")
Text(0.5, 0, 'wavenumber (cm-1)')
That’s it!