Filtering Lines by Quantum Number
In this tutorial, we filter lines by the difference of vibrational quantum number between lower and upper states, \(\Delta \nu\).
from exojax.utils.grids import wavenumber_grid
nu_grid, wav, resolution = wavenumber_grid(24000.0, 26000,80000, unit="AA", xsmode="premodit")
xsmode = premodit
xsmode assumes ESLOG in wavenumber space: mode=premodit
Normally, “mdb” performs the activation automatically. We would like to avoid this automatic activation and perform activation after the user defines the mask. To do this, call mdb with activation=False. We also would use the quantum number available in this database. So, use optional_quantum_states=True.
from exojax.database.exomol.api import MdbExomol
mdb =MdbExomol("CO/12C-16O/Li2015/", nu_grid, optional_quantum_states=True, activation=False)
HITRAN exact name= (12C)(16O)
Background atmosphere: H2
Reading CO/12C-16O/Li2015/12C-16O__Li2015.trans.bz2
DataFrame (self.df) available.
When called activation turn off, the DataFrame becomes automatically available. Check mdb.df.
print(mdb.df)
# i_upper i_lower A nu_lines gup jlower jupper elower v_l v_u kp_l kp_u Sij0
0 84 42 1.155e-06 2.405586 3 0 1 66960.7124 41 41 e e 3.811968898414225e-164
1 83 41 1.161e-06 2.441775 3 0 1 65819.903 40 40 e e 9.663028103692631e-162
2 82 40 1.162e-06 2.477774 3 0 1 64654.9206 39 39 e e 2.7438392479197905e-159
3 81 39 1.159e-06 2.513606 3 0 1 63465.8042 38 38 e e 8.73322833971394e-157
4 80 38 1.152e-06 2.549292 3 0 1 62252.5793 37 37 e e 3.115220404216648e-154
... ... ... ... ... ... ... ... ... ... ... ... ... ...
125,491 306 253 7.164e-10 22147.135424 15 6 7 80.7354 0 11 e e 1.8282485593637477e-31
125,492 474 421 9.852e-10 22147.86595 23 10 11 211.4041 0 11 e e 2.0425455665383687e-31
125,493 348 295 7.72e-10 22147.897299 17 7 8 107.6424 0 11 e e 1.9589545250222689e-31
125,494 432 379 9.056e-10 22148.262711 21 9 10 172.978 0 11 e e 2.0662209116961706e-31
125,495 390 337 8.348e-10 22148.273111 19 8 9 138.3903 0 11 e e 2.0387827253771594e-31
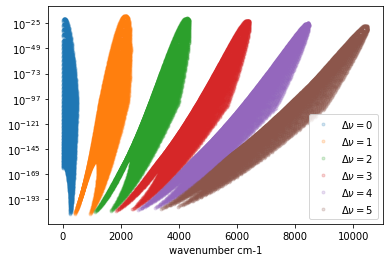
So, we can use the above information for masking the molecular lines. DataFrame is not limited by nu_grids. So, all of the lines are visible (but using lazy I/O of vaex). Let’s plot the line strength with different \(\Delta \nu\).
import matplotlib.pyplot as plt
for dv in range(0, 6):
mask = mdb.df["v_u"] - mdb.df["v_l"] == dv
dfv = mdb.df[mask]
plt.plot(dfv["nu_lines"].values,
dfv["Sij0"].values,
".",
label="$\\Delta \\nu = $" + str(dv),
alpha=0.2)
plt.legend()
plt.yscale("log")
plt.xlabel("wavenumber cm-1")
plt.show()
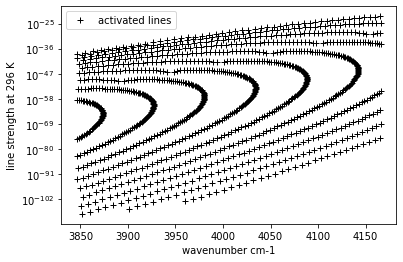
In ExoJAX, we need activate “mdb”. User-defined mask for DataFrame can be used as load_mask option of msb.activate.
load_mask = (mdb.df["v_u"] - mdb.df["v_l"] == 2)
mdb.activate(mdb.df, load_mask)
.broad is used.
Broadening code level= a0
default broadening parameters are used for 69 J lower states in 150 states
The following lines are activated and can be used further analysis as usual.
plt.plot(mdb.nu_lines,
mdb.line_strength_ref,
"+",
color="black",
label="activated lines")
plt.legend()
plt.ylabel("line strength at 296 K")
plt.xlabel("wavenumber cm-1")
plt.yscale("log")
plt.show()
# %%
Note that the above process is also applicable to MdbHitemp and MdbHitran.
In tests/integration/moldb these examples are available for reference.
quantum_states_filter_exomol.py
quantum_states_filter_hitemp.py
quantum_states_filter_hitran_co.py