Line identification using ExoMolHR
Hajime Kawahara 5/16 (2025)
ExoMolHR is an empirical, high-resolution molecular spectrum calculator for the high-temperature molecular line lists available from the ExoMol molecular database.
ExoMolHR provides an API that delivers selected line information from ExoMol when a temperature and wavenumber range are specified. This is particularly useful in cases where it is unnecessary to use the complete but large ExoMol database directly. ExoJAX is capable of handling ExoMolHR. Here, let us use ExoMolHR from the perspective of line identification, to examine which strong lines are present at a given temperature.
To first check which molecules are available, use the following function:
from exojax.database.exomolhr.api import list_exomolhr_molecules
molecules = list_exomolhr_molecules()
print(molecules)
['AlCl', 'AlH', 'AlO', 'C2', 'C2H2', 'CaH', 'CH4', 'CN', 'CO2', 'H2CO', 'H2O', 'H2S', 'H3+', 'H3O+', 'LiOH', 'MgH', 'NH', 'NH3', 'NO', 'SiN', 'SiO', 'SO', 'SO2', 'TiO', 'YO', 'ZrO', 'BeH', 'CaOH', 'H2CS', 'N2O', 'OCS', 'PN', 'VO']
To find the available isotopologues for each molecule, use the following function:
from exojax.database.exomolhr.api import list_exomolhr_isotopes
iso_dict = list_exomolhr_isotopes(molecules)
print(iso_dict)
{'CaH': ['40Ca-1H'], 'H3+': ['1H2-2H_p', '1H3_p', '2H2-1H_p', '2H3_p'], 'AlH': ['27Al-1H'], 'LiOH': ['6Li-16O-1H', '7Li-16O-1H'], 'C2': ['12C2'], 'C2H2': ['12C2-1H2'], 'H2CO': ['1H2-12C-16O'], 'CN': ['12C-14N'], 'CH4': ['12C-1H4'], 'CO2': ['12C-16O2'], 'MgH': ['24Mg-1H', '25Mg-1H', '26Mg-1H'], 'H2S': ['1H2-32S'], 'H2O': ['1H2-16O'], 'AlCl': ['27Al-35Cl', '27Al-37Cl'], 'H3O+': ['1H3-16O_p'], 'AlO': ['26Al-16O', '27Al-16O', '27Al-17O', '27Al-18O'], 'NH': ['14N-1H', '14N-2H', '15N-1H', '15N-2H'], 'NH3': ['14N-1H3', '15N-1H3'], 'SiN': ['28Si-14N', '28Si-15N', '29Si-14N', '30Si-14N'], 'SO2': ['32S-16O2'], 'ZrO': ['90Zr-16O', '91Zr-16O', '92Zr-16O', '93Zr-16O', '94Zr-16O', '96Zr-16O'], 'SO': ['32S-16O'], 'SiO': ['28Si-16O'], 'NO': ['14N-16O'], 'TiO': ['48Ti-16O'], 'BeH': ['9Be-1H', '9Be-2H'], 'H2CS': ['1H2-12C-32S'], 'YO': ['89Y-16O', '89Y-17O', '89Y-18O'], 'N2O': ['14N2-16O'], 'CaOH': ['40Ca-16O-1H'], 'PN': ['31P-14N', '31P-15N'], 'OCS': ['16O-12C-32S'], 'VO': ['51V-16O']}
To load a molecular database, you can use XdbExomolHR. The Xdb refers to an extra database outside of Mdb, Adb, Cdb, and Pdb, and each of these has its own dedicated interface.
from exojax.database import XdbExomolHR
from exojax.utils.grids import wavenumber_grid
import matplotlib.pyplot as plt
nus, _, _ = wavenumber_grid(22800.0, 23600.0, 10, xsmode="premodit", unit="AA")
temperature = 1300.0
iso = "1H2-16O"
xdb = XdbExomolHR(iso, nus, temperature)
xsmode = premodit xsmode assumes ESLOG in wavenumber space: xsmode=premodit Your wavelength grid is in * descending * order The wavenumber grid is in ascending order by definition. Please be careful when you use the wavelength grid. HITRAN exact name= H2(16O)
/home/kawahara/exojax/src/exojax/utils/grids.py:85: UserWarning: Both input wavelength and output wavenumber are in ascending order.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/grids.py:249: UserWarning: Resolution may be too small. R=260.97413588061954
warnings.warn("Resolution may be too small. R=" + str(resolution), UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
Downloaded and unzipped to opacity_zips/20250806073433__1H2-16O__1300.0K.csv
XdbExomolHR shares several common attributes with MdbExomol.
However, unlike MdbExomol, it does not support changing the
temperature or provide information such as the partition function.
xdb.line_strength, xdb.jlower, xdb.elower
(array([2.07935666e-33, 1.00589019e-32, 1.89912653e-36, ...,
2.28603692e-35, 3.74997478e-32, 2.22317669e-32]),
array([ 7, 9, 10, ..., 14, 7, 7]),
array([13576.566165, 9837.808903, 13685.911191, ..., 5940.635876,
7350.151429, 14753.618734]))
Now, let us get xdb at a given temperature over a specified
wavelength range for all isotopologues available in ExoMolHR.
k=0
xdbs = {}
for molecule in iso_dict:
isos = iso_dict[molecule]
for j, iso in enumerate(isos):
try:
xdb = XdbExomolHR(iso, nus, temperature, crit=1.e-24)
xdbs[iso] = xdb
except:
k=k+1
print(f"No line? {iso}")
print(k, "molecules have no lines")
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
HITRAN exact name= (40Ca)H
HITRAN exact name= (40Ca)H
Downloaded and unzipped to opacity_zips/20250806073456__40Ca-1H__1300.0K.csv
HITRAN exact name= H2(2H_p)
HITRAN exact name= H2(2H_p)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073458__1H2-2H_p__1300.0K.csv
HITRAN exact name= (1H3_p)
HITRAN exact name= (1H3_p)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073500__1H3_p__1300.0K.csv
HITRAN exact name= D2(1H_p)
HITRAN exact name= D2(1H_p)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073502__2H2-1H_p__1300.0K.csv
HITRAN exact name= (2H3_p)
HITRAN exact name= (2H3_p)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073504__2H3_p__1300.0K.csv
No line? 2H3_p
HITRAN exact name= (27Al)H
HITRAN exact name= (27Al)H
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073506__27Al-1H__1300.0K.csv
HITRAN exact name= (6Li)(16O)H
HITRAN exact name= (6Li)(16O)H
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073508__6Li-16O-1H__1300.0K.csv
No line? 6Li-16O-1H
HITRAN exact name= (7Li)(16O)H
HITRAN exact name= (7Li)(16O)H
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073509__7Li-16O-1H__1300.0K.csv
No line? 7Li-16O-1H
HITRAN exact name= (12C)2
HITRAN exact name= (12C)2
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073511__12C2__1300.0K.csv
HITRAN exact name= (12C)2H2
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
Downloaded and unzipped to opacity_zips/20250806073514__12C2-1H2__1300.0K.csv
No line? 12C2-1H2
HITRAN exact name= H2(12C)(16O)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
Downloaded and unzipped to opacity_zips/20250806073517__1H2-12C-16O__1300.0K.csv
HITRAN exact name= (12C)(14N)
HITRAN exact name= (12C)(14N)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073519__12C-14N__1300.0K.csv
HITRAN exact name= (12C)(1H)4
HITRAN exact name= (12C)(1H)4
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073522__12C-1H4__1300.0K.csv
HITRAN exact name= (12C)(16O)2
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
Downloaded and unzipped to opacity_zips/20250806073535__12C-16O2__1300.0K.csv
No line? 12C-16O2
HITRAN exact name= (24Mg)H
HITRAN exact name= (24Mg)H
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073540__24Mg-1H__1300.0K.csv
No line? 24Mg-1H
HITRAN exact name= (25Mg)H
HITRAN exact name= (25Mg)H
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073542__25Mg-1H__1300.0K.csv
HITRAN exact name= (26Mg)H
HITRAN exact name= (26Mg)H
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073544__26Mg-1H__1300.0K.csv
HITRAN exact name= H2(32S)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
Downloaded and unzipped to opacity_zips/20250806073546__1H2-32S__1300.0K.csv
HITRAN exact name= H2(16O)
Downloaded and unzipped to opacity_zips/20250806073433__1H2-16O__1300.0K.csv
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
HITRAN exact name= (27Al)(35Cl)
HITRAN exact name= (27Al)(35Cl)
Downloaded and unzipped to opacity_zips/20250806073550__27Al-35Cl__1300.0K.csv
No line? 27Al-35Cl
HITRAN exact name= (27Al)(37Cl)
HITRAN exact name= (27Al)(37Cl)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073552__27Al-37Cl__1300.0K.csv
No line? 27Al-37Cl
HITRAN exact name= (1H)3(16O_p)
HITRAN exact name= (1H)3(16O_p)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073553__1H3-16O_p__1300.0K.csv
No line? 1H3-16O_p
HITRAN exact name= (26Al)(16O)
HITRAN exact name= (26Al)(16O)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073555__26Al-16O__1300.0K.csv
HITRAN exact name= (27Al)(16O)
HITRAN exact name= (27Al)(16O)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073558__27Al-16O__1300.0K.csv
HITRAN exact name= (27Al)(17O)
HITRAN exact name= (27Al)(17O)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073600__27Al-17O__1300.0K.csv
HITRAN exact name= (27Al)(18O)
HITRAN exact name= (27Al)(18O)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073603__27Al-18O__1300.0K.csv
HITRAN exact name= (14N)H
HITRAN exact name= (14N)H
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073605__14N-1H__1300.0K.csv
No line? 14N-1H
HITRAN exact name= (14N)D
HITRAN exact name= (14N)D
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073607__14N-2H__1300.0K.csv
HITRAN exact name= (15N)H
HITRAN exact name= (15N)H
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073609__15N-1H__1300.0K.csv
No line? 15N-1H
HITRAN exact name= (15N)D
HITRAN exact name= (15N)D
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073611__15N-2H__1300.0K.csv
HITRAN exact name= (14N)(1H)3
HITRAN exact name= (14N)(1H)3
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073613__14N-1H3__1300.0K.csv
HITRAN exact name= (15N)(1H)3
HITRAN exact name= (15N)(1H)3
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073616__15N-1H3__1300.0K.csv
HITRAN exact name= (28Si)(14N)
HITRAN exact name= (28Si)(14N)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073618__28Si-14N__1300.0K.csv
No line? 28Si-14N
HITRAN exact name= (28Si)(15N)
HITRAN exact name= (28Si)(15N)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073620__28Si-15N__1300.0K.csv
No line? 28Si-15N
HITRAN exact name= (29Si)(14N)
HITRAN exact name= (29Si)(14N)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073622__29Si-14N__1300.0K.csv
No line? 29Si-14N
HITRAN exact name= (30Si)(14N)
HITRAN exact name= (30Si)(14N)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073624__30Si-14N__1300.0K.csv
No line? 30Si-14N
HITRAN exact name= (32S)(16O)2
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
Downloaded and unzipped to opacity_zips/20250806073626__32S-16O2__1300.0K.csv
No line? 32S-16O2
HITRAN exact name= (90Zr)(16O)
HITRAN exact name= (90Zr)(16O)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073629__90Zr-16O__1300.0K.csv
HITRAN exact name= (91Zr)(16O)
HITRAN exact name= (91Zr)(16O)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073631__91Zr-16O__1300.0K.csv
HITRAN exact name= (92Zr)(16O)
HITRAN exact name= (92Zr)(16O)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073633__92Zr-16O__1300.0K.csv
HITRAN exact name= (93Zr)(16O)
HITRAN exact name= (93Zr)(16O)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073635__93Zr-16O__1300.0K.csv
HITRAN exact name= (94Zr)(16O)
HITRAN exact name= (94Zr)(16O)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073637__94Zr-16O__1300.0K.csv
HITRAN exact name= (96Zr)(16O)
HITRAN exact name= (96Zr)(16O)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073638__96Zr-16O__1300.0K.csv
HITRAN exact name= (32S)(16O)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
Downloaded and unzipped to opacity_zips/20250806073640__32S-16O__1300.0K.csv
No line? 32S-16O
HITRAN exact name= (28Si)(16O)
HITRAN exact name= (28Si)(16O)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073642__28Si-16O__1300.0K.csv
No line? 28Si-16O
HITRAN exact name= (14N)(16O)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
Downloaded and unzipped to opacity_zips/20250806073644__14N-16O__1300.0K.csv
No line? 14N-16O
HITRAN exact name= (48Ti)(16O)
HITRAN exact name= (48Ti)(16O)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073646__48Ti-16O__1300.0K.csv
No line? 48Ti-16O
HITRAN exact name= (9Be)H
HITRAN exact name= (9Be)H
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073648__9Be-1H__1300.0K.csv
No line? 9Be-1H
HITRAN exact name= (9Be)D
HITRAN exact name= (9Be)D
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073650__9Be-2H__1300.0K.csv
HITRAN exact name= H2(12C)(32S)
HITRAN exact name= H2(12C)(32S)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073652__1H2-12C-32S__1300.0K.csv
No line? 1H2-12C-32S
HITRAN exact name= (89Y)(16O)
HITRAN exact name= (89Y)(16O)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073654__89Y-16O__1300.0K.csv
No line? 89Y-16O
HITRAN exact name= (89Y)(17O)
HITRAN exact name= (89Y)(17O)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073656__89Y-17O__1300.0K.csv
No line? 89Y-17O
HITRAN exact name= (89Y)(18O)
HITRAN exact name= (89Y)(18O)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073658__89Y-18O__1300.0K.csv
No line? 89Y-18O
HITRAN exact name= (14N)2(16O)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
Downloaded and unzipped to opacity_zips/20250806073700__14N2-16O__1300.0K.csv
HITRAN exact name= (40Ca)(16O)H
HITRAN exact name= (40Ca)(16O)H
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073707__40Ca-16O-1H__1300.0K.csv
No line? 40Ca-16O-1H
HITRAN exact name= (31P)(14N)
HITRAN exact name= (31P)(14N)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073709__31P-14N__1300.0K.csv
No line? 31P-14N
HITRAN exact name= (31P)(15N)
HITRAN exact name= (31P)(15N)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
No line? 31P-15N
HITRAN exact name= (16O)(12C)(32S)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
Downloaded and unzipped to opacity_zips/20250806073713__16O-12C-32S__1300.0K.csv
No line? 16O-12C-32S
HITRAN exact name= (51V)(16O)
HITRAN exact name= (51V)(16O)
/home/kawahara/exojax/src/exojax/utils/molname.py:197: FutureWarning: e2s will be replaced to exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/utils/molname.py:63: UserWarning: No isotope number identified.
warnings.warn("No isotope number identified.", UserWarning)
/home/kawahara/exojax/src/exojax/utils/molname.py:91: FutureWarning: exojax.utils.molname.exact_molname_exomol_to_simple_molname will be replaced to radis.api.exomolapi.exact_molname_exomol_to_simple_molname.
warnings.warn(
/home/kawahara/exojax/src/exojax/database/molinfo.py:38: UserWarning: exact molecule name is not Exomol nor HITRAN form.
warnings.warn("exact molecule name is not Exomol nor HITRAN form.")
/home/kawahara/exojax/src/exojax/database/molinfo.py:39: UserWarning: No molmass available
warnings.warn("No molmass available", UserWarning)
Downloaded and unzipped to opacity_zips/20250806073715__51V-16O__1300.0K.csv
No line? 51V-16O
30 molecules have no lines
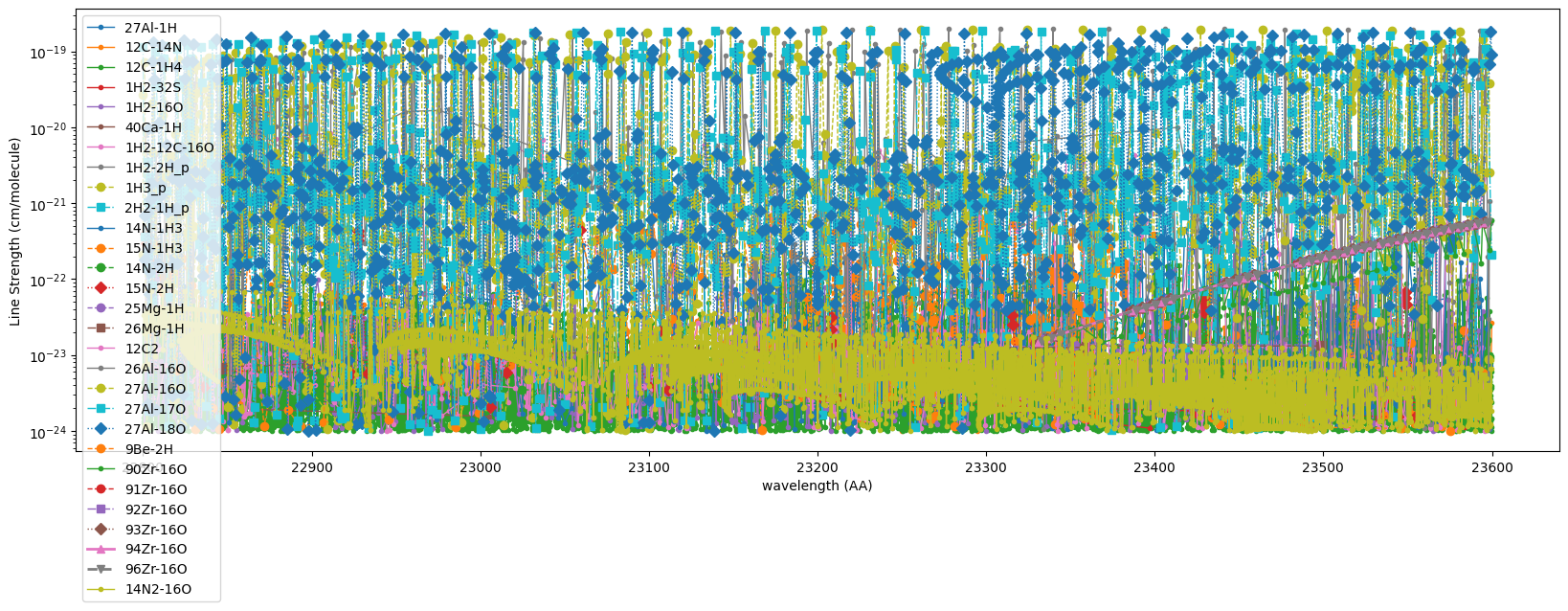
Let’s plot the line strength in this wavelength region. You can check which isotope has strong lines.
lslist = ["-", "--", "-.", ":", "-", "--", "-.", ":"]
lwlist = [1.0, 1.0, 1.0, 1.0, 2.0, 2.0, 2.0, 2.0]
markers_list = [".", "o", "s", "D", "^", "v", "<", ">"]
fig = plt.figure(figsize=(20, 6))
for molecule in iso_dict:
isos = iso_dict[molecule]
for j, iso in enumerate(isos):
try:
xdb = xdbs[iso]
plt.plot(1.e8/xdb.nu_lines, xdb.line_strength, markers_list[j], label=iso, ls=lslist[j], lw=lwlist[j])
except:
print(f"No line? {iso}")
continue
plt.yscale("log")
plt.xlabel("wavelength (AA)")
plt.ylabel("Line Strength (cm/molecule)")
plt.legend()
plt.show()
No line? 2H3_p
No line? 6Li-16O-1H
No line? 7Li-16O-1H
No line? 12C2-1H2
No line? 12C-16O2
No line? 24Mg-1H
No line? 27Al-35Cl
No line? 27Al-37Cl
No line? 1H3-16O_p
No line? 14N-1H
No line? 15N-1H
No line? 28Si-14N
No line? 28Si-15N
No line? 29Si-14N
No line? 30Si-14N
No line? 32S-16O2
No line? 32S-16O
No line? 28Si-16O
No line? 14N-16O
No line? 48Ti-16O
No line? 9Be-1H
No line? 1H2-12C-32S
No line? 89Y-16O
No line? 89Y-17O
No line? 89Y-18O
No line? 40Ca-16O-1H
No line? 31P-14N
No line? 31P-15N
No line? 16O-12C-32S
No line? 51V-16O
that’s it.