Cross Section for Many Lines using MODIT
Update: Febrary 7th/2025, Hajime Kawahara
We demonstarte the Modified Discrete Integral Transform (MODIT), which is the modified version of DIT for exojax. MODIT uses the evenly-spaced logarithm grid (ESLOG) as a wavenumber dimension. MODIT takes advantage especially for the case that the number of the molecular line is large (typically > 10,000). We here compare the results by MODIT with the direct computation (LPF).
Here, we use FP64, but if you want you can use FP32 (but slightly large errors):
from jax import config
config.update("jax_enable_x64", True)
import matplotlib.pyplot as plt
from exojax.database.core.line_strength import line_strength, doppler_sigma, gamma_hitran, gamma_natural
from exojax.database.exomol.api import MdbExomol
from exojax.utils.grids import wavenumber_grid
from exojax.utils.constants import Tref_original
# Setting wavenumber bins and loading HITRAN database
nus, wav, R = wavenumber_grid(1900.0, 2300.0, 350000, unit="cm-1", xsmode="modit")
mdbCO = MdbHitran("CO", nus, isotope=1) # use isotope=1 12C-16O
# set T, P and partition function
Mmol = mdbCO.molmass
Tfix = 1000.0 # we assume T=1000K
Pfix = 1.0e-3 # we compute P=1.e-3 bar
Ppart = Pfix # partial pressure of CO. here we assume a 100% CO atmosphere
xsmode = modit xsmode assumes ESLOG in wavenumber space: xsmode=modit ====================================================================== The wavenumber grid should be in ascending order. The users can specify the order of the wavelength grid by themselves. Your wavelength grid is in * descending * order ====================================================================== radis engine = vaex
qt = mdbCO.qr_interp(1, Tfix, Tref_original) # isotope=1
# computes logsij0 etc in device
mdbCO.generate_jnp_arrays()
# compute Sij, gamma_L, sigmaD
Sij = line_strength(
Tfix, mdbCO.logsij0, mdbCO.nu_lines, mdbCO.elower, qt, Tref_original
)
gammaL = gamma_hitran(
Pfix, Tfix, Ppart, mdbCO.n_air, mdbCO.gamma_air, mdbCO.gamma_self
) + gamma_natural(mdbCO.A)
MODIT uses the normalized quantities by wavenumber/R, where R is the spectral resolution. In this case, the normalized Doppler width (nsigmaD) is common for the same isotope. Then, we use a 2D DIT grid with the normalized gammaL and q = R log(nu).
from exojax.database.hitran import normalized_doppler_sigma
dv_lines = mdbCO.nu_lines / R
nsigmaD = normalized_doppler_sigma(Tfix, Mmol, R)
ngammaL = gammaL / dv_lines
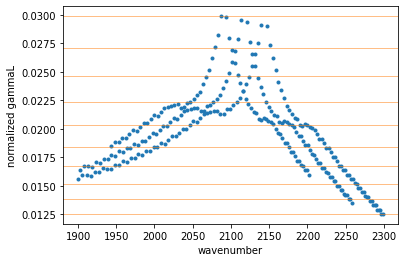
MODIT uses a grid of ngammaL, and wavenumber. set_ditgrid.ditgrid_log_interval makes a 1D grid (evenly log spaced) for ngamma.
from exojax.opacity._common.set_ditgrid import ditgrid_log_interval
ngammaL_grid = ditgrid_log_interval(ngammaL)
# show the grids
plt.plot(mdbCO.nu_lines, ngammaL, ".")
for i in ngammaL_grid:
plt.axhline(i, lw=1, alpha=0.5, color="C1")
plt.xlabel("wavenumber")
plt.ylabel("normalized gammaL")
Text(0, 0.5, 'normalized gammaL')
We need to precompute the contribution for wavenumber and pmarray. These can be computed using init_dit.
from exojax.opacity import initspec
cnu, indexnu, R, pmarray = initspec.init_modit(mdbCO.nu_lines, nus)
Let’s compute the cross section!
from exojax.opacity.modit.modit_scanfft import xsvector_scanfft
xs = xsvector_scanfft(cnu, indexnu, R, pmarray, nsigmaD, ngammaL, Sij, nus, ngammaL_grid)
Also, we here try the direct computation using LPF for the comparison purpose
from exojax.opacity import OpaDirect
opa = OpaDirect(mdbCO, nus)
xsv = opa.xsvector(Tfix, Pfix, Ppart)
fig = plt.figure(figsize=(10, 5))
ax = fig.add_subplot(211)
plt.plot(nus, xs, lw=1, alpha=0.5, label="MODIT")
plt.plot(nus, xsv, lw=1, alpha=0.5, label="Direct LPF")
plt.legend(loc="upper right")
plt.ylabel("Cross Section (cm2)")
ax = fig.add_subplot(212)
plt.plot(nus, xsv - xs, lw=2, alpha=0.5, label="MODIT")
plt.ylabel("LPF - DIT (cm2)")
plt.legend(loc="upper left")
plt.show()
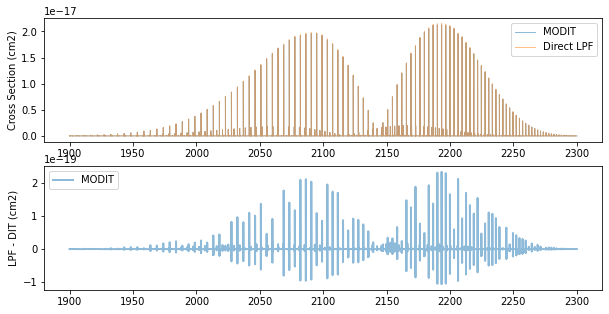
There is about 1 % deviation between LPF and MODIT.
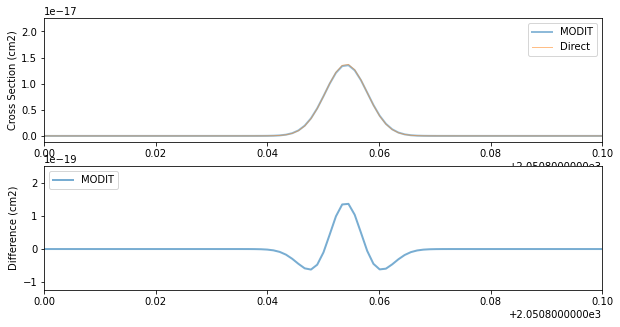
fig = plt.figure(figsize=(10, 5))
ax = fig.add_subplot(211)
plt.plot(nus, xs, lw=2, alpha=0.5, label="MODIT")
plt.plot(nus, xsv, lw=1, alpha=0.5, label="Direct")
plt.legend(loc="upper right")
plt.xlim(2050.8, 2050.9)
plt.ylabel("Cross Section (cm2)")
ax = fig.add_subplot(212)
plt.plot(nus, xsv - xs, lw=2, alpha=0.6, label="MODIT")
plt.legend(loc="upper left")
plt.ylabel("Difference (cm2)")
plt.xlim(2050.8, 2050.9)
# plt.yscale("log")
plt.savefig("fine_grid.png")