Forward Modeling of an Emission Spectrum¶
from exojax.spec import rtransfer as rt
#ATMOSPHERE
NP=100
T0=1295.0 #K
Parr, dParr, k=rt.pressure_layer(NP=NP)
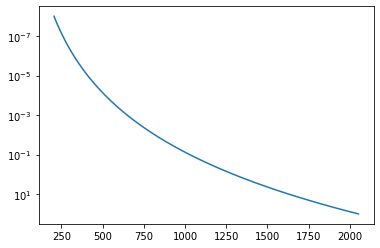
Tarr = T0*(Parr)**0.1
A T-P profile we assume is …
import matplotlib.pyplot as plt
plt.plot(Tarr,Parr)
plt.yscale("log")
plt.gca().invert_yaxis()
plt.show()
We set a wavenumber grid using wavenumber_grid.
from exojax.utils.grids import wavenumber_grid
nus,wav,res=wavenumber_grid(22920,23000,1000,unit="AA")
xsmode assumes ESLOG in wavenumber space: mode=lpf
/home/kawahara/exojax/src/exojax/utils/grids.py:124: UserWarning: Resolution may be too small. R=286712.70993002696
warnings.warn('Resolution may be too small. R=' + str(resolution),
Loading a molecular database of CO and CIA (H2-H2)…
from exojax.spec import api, contdb
mdbCO=api.MdbExomol('.database/CO/12C-16O/Li2015',nus,crit=1.e-46)
cdbH2H2=contdb.CdbCIA('.database/H2-H2_2011.cia',nus)
Background atmosphere: H2
Reading .database/CO/12C-16O/Li2015/12C-16O__Li2015.trans.bz2
.broad is used.
Broadening code level= a0
H2-H2
from exojax.spec import molinfo
molmassCO=molinfo.molmass("CO")
Computing the relative partition function,
from jax import vmap
qt=vmap(mdbCO.qr_interp)(Tarr)
Pressure and Natural broadenings
from jax import jit
from exojax.spec.exomol import gamma_exomol
from exojax.spec import gamma_natural
gammaLMP = jit(vmap(gamma_exomol,(0,0,None,None)))\
(Parr,Tarr,mdbCO.n_Texp,mdbCO.alpha_ref)
gammaLMN=gamma_natural(mdbCO.A)
gammaLM=gammaLMP+gammaLMN[None,:]
Doppler broadening
from exojax.spec import doppler_sigma
sigmaDM=jit(vmap(doppler_sigma,(None,0,None)))\
(mdbCO.nu_lines,Tarr,molmassCO)
And line strength
from exojax.spec import SijT
SijM=jit(vmap(SijT,(0,None,None,None,0)))\
(Tarr,mdbCO.logsij0,mdbCO.nu_lines,mdbCO.elower,qt)
nu matrix
from exojax.spec import make_numatrix0
numatrix=make_numatrix0(nus,mdbCO.nu_lines)
Or you can use initspec.init_lpf instead.
#Or you can use initspec.init_lpf instead.
from exojax.spec import initspec
numatrix=initspec.init_lpf(mdbCO.nu_lines,nus)
Providing numatrix, thermal broadening, gamma, and line strength, we can compute cross section.
from exojax.spec.lpf import xsmatrix
xsm=xsmatrix(numatrix,sigmaDM,gammaLM,SijM)
xsmatrix has the shape of (# of layers, # of nu grid)
import numpy as np
np.shape(xsm)
(100, 1000)
import numpy as np
plt.imshow(xsm,cmap="afmhot")
plt.show()

computing delta tau for CO
from exojax.spec.rtransfer import dtauM
Rp=0.88
Mp=33.2
g=2478.57730044555*Mp/Rp**2
#g=1.e5 #gravity cm/s2
MMR=0.0059 #mass mixing ratio
dtaum=dtauM(dParr,xsm,MMR*np.ones_like(Tarr),molmassCO,g)
computing delta tau for CIA
from exojax.spec.rtransfer import dtauCIA
mmw=2.33 #mean molecular weight
mmrH2=0.74
molmassH2=molinfo.molmass("H2")
vmrH2=(mmrH2*mmw/molmassH2) #VMR
dtaucH2H2=dtauCIA(nus,Tarr,Parr,dParr,vmrH2,vmrH2,\
mmw,g,cdbH2H2.nucia,cdbH2H2.tcia,cdbH2H2.logac)
The total delta tau is a summation of them
dtau=dtaum+dtaucH2H2
you can plot a contribution function using exojax.plot.atmplot
from exojax.plot.atmplot import plotcf
plotcf(nus,dtau,Tarr,Parr,dParr)
plt.show()

radiative transfering…
from exojax.spec import planck
from exojax.spec.rtransfer import rtrun
sourcef = planck.piBarr(Tarr,nus)
F0=rtrun(dtau,sourcef)
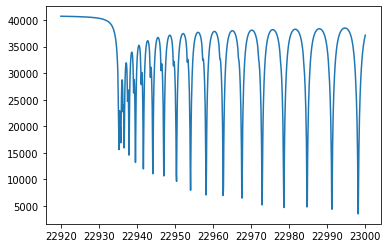
plt.plot(wav[::-1],F0)
[<matplotlib.lines.Line2D at 0x7f2baa2c2970>]
applying an instrumental response and planet/stellar rotation to the raw spectrum
from exojax.spec import response
from exojax.utils.constants import c
import jax.numpy as jnp
wavd=jnp.linspace(22920,23000,500) #observational wavelength grid
nusd = 1.e8/wavd[::-1]
RV=10.0 #RV km/s
vsini=20.0 #Vsini km/s
u1=0.0 #limb darkening u1
u2=0.0 #limb darkening u2
R=100000.
beta=c/(2.0*np.sqrt(2.0*np.log(2.0))*R) #IP sigma need check
Frot=response.rigidrot(nus,F0,vsini,u1,u2)
F=response.ipgauss_sampling(nusd,nus,Frot,beta,RV)
plt.plot(wav[::-1],F0)
plt.plot(wavd[::-1],F)
[<matplotlib.lines.Line2D at 0x7f2baa4ff190>]
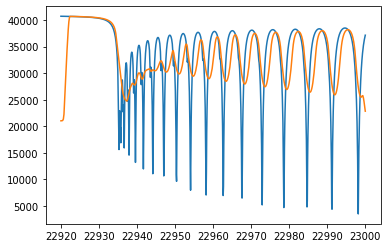
The flux decreases at the edges of the left and right sides are artificial due to the convolution. You might need to some margins of the wavenumber range to eliminate these artifacts.
np.savetxt("spectrum.txt",np.array([wavd,F]).T,delimiter=",")